System for multi-round editing of fungal genome by CRISPR system and method thereof
A gene editing and genome technology, applied in the field of genetic engineering and biology, can solve the problems of fewer types of resistance markers, lower positive rate, and difficult gene editing tasks
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
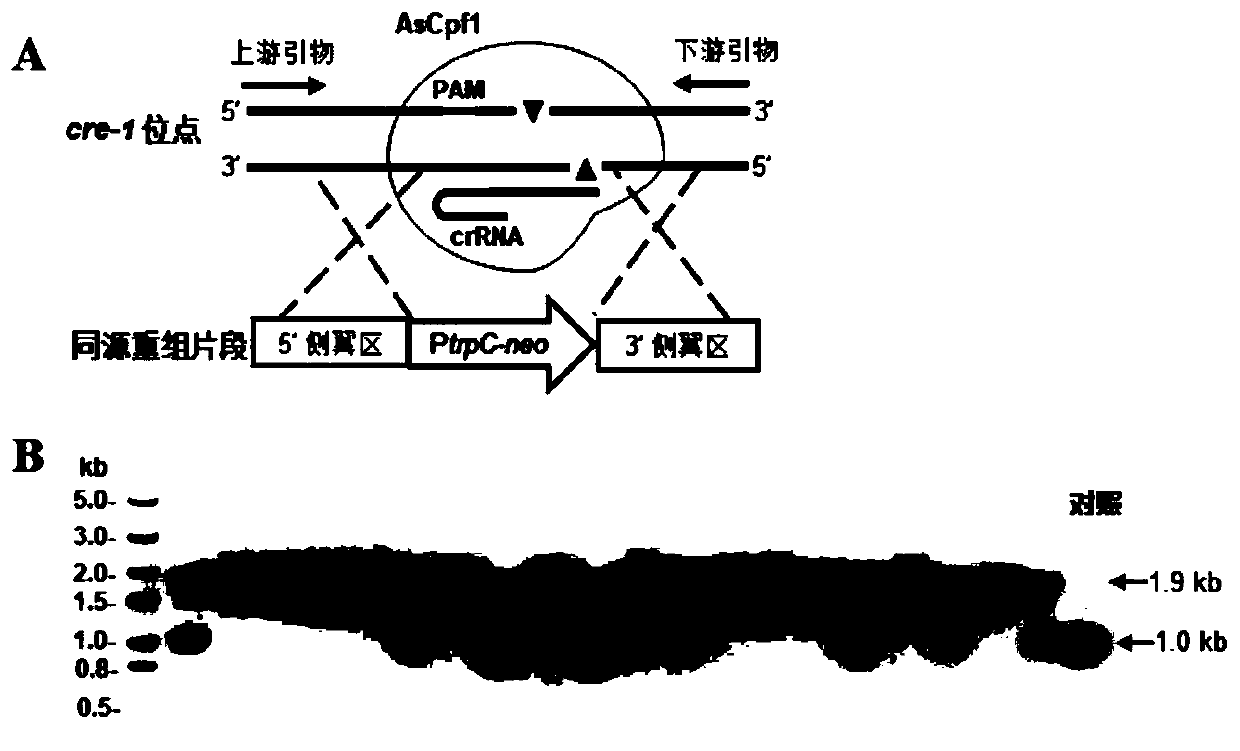
[0045]Example 1. Construction of the Myceliophthora thermophila genome editing system mediated by CRISPR-Cas12a (AsCpf1)
[0046] 1. Construction of Cas12a (AsCpf1) expression cassette vector
[0047] With plasmid p0380-bar (Liu Q, Gao RR, Li JG, Lin LC, Zhao JQ, Sun WL, TianCG. Development of a genome-editing CRISPR / Cas9system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase productionstrain engineering. Biotechnology for Biofuels.2017, 10:1.) to construct an expression vector for the backbone. With reference to the genome of Myceliophthora thermophila, Cas12a (AsCpf1) is selected from the AsCpf1 protein of Acidaminococcussp.BV3L6 to carry out codon bias optimization (codons are optimized according to the expression preference of Myceliophthora thermophila), in the N of AsCpf1 protein The nuclear localization signal sequence (PPRKRAKTEDE) of Myceliophthora thermophila transcription factor hacI (MYCTH_2310995) was added to the C-terminal an...
Embodiment 2
[0082] Example 2, CRISPR-Cas12a (AsCpf1) system for simultaneous editing of multiple genes in the Myceliophthora thermophila genome
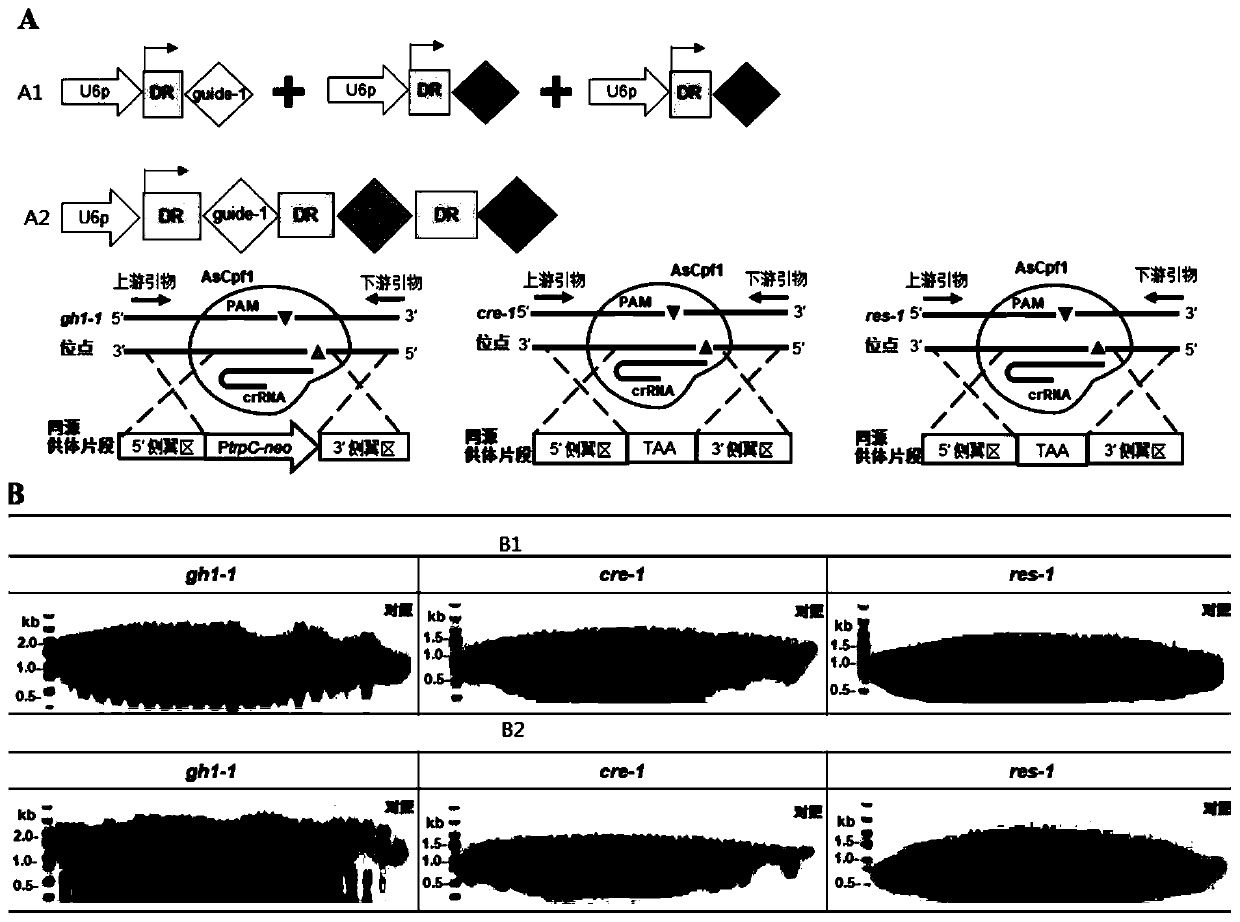
[0083] 1. The crRNA single-plasmid system realizes editing of multiple genes in the genome
[0084] The first group of editing system is to mix Cas enzyme expression box Ptef1-AsCpf1-TtprC, crRNA expression elements U6p-crRNA-cre1, U6p-crRNA-res1 and U6p-crRNA-gh1-1 in molar ratio, and its homologous supply The body DNA fragments donor-2-cre1, donor-res1 and donor-1-gh1-1 are mixed in equimolecular molar ratios, the above three are the total amount of Cas enzyme expression box, crRNA and homologous donor DNA in molecular After co-transformation into the protoplast cells of Myceliophthora thermophila wild-type strain ATCC 42464 at a molar ratio of 1:1:1, Cas12a (AsCpf1) was mediated by crRNA through the protospacer and the DNA of the target gene on the genome of the host cell The strands are paired to recognize the target site for cleavage, and ...
Embodiment 3
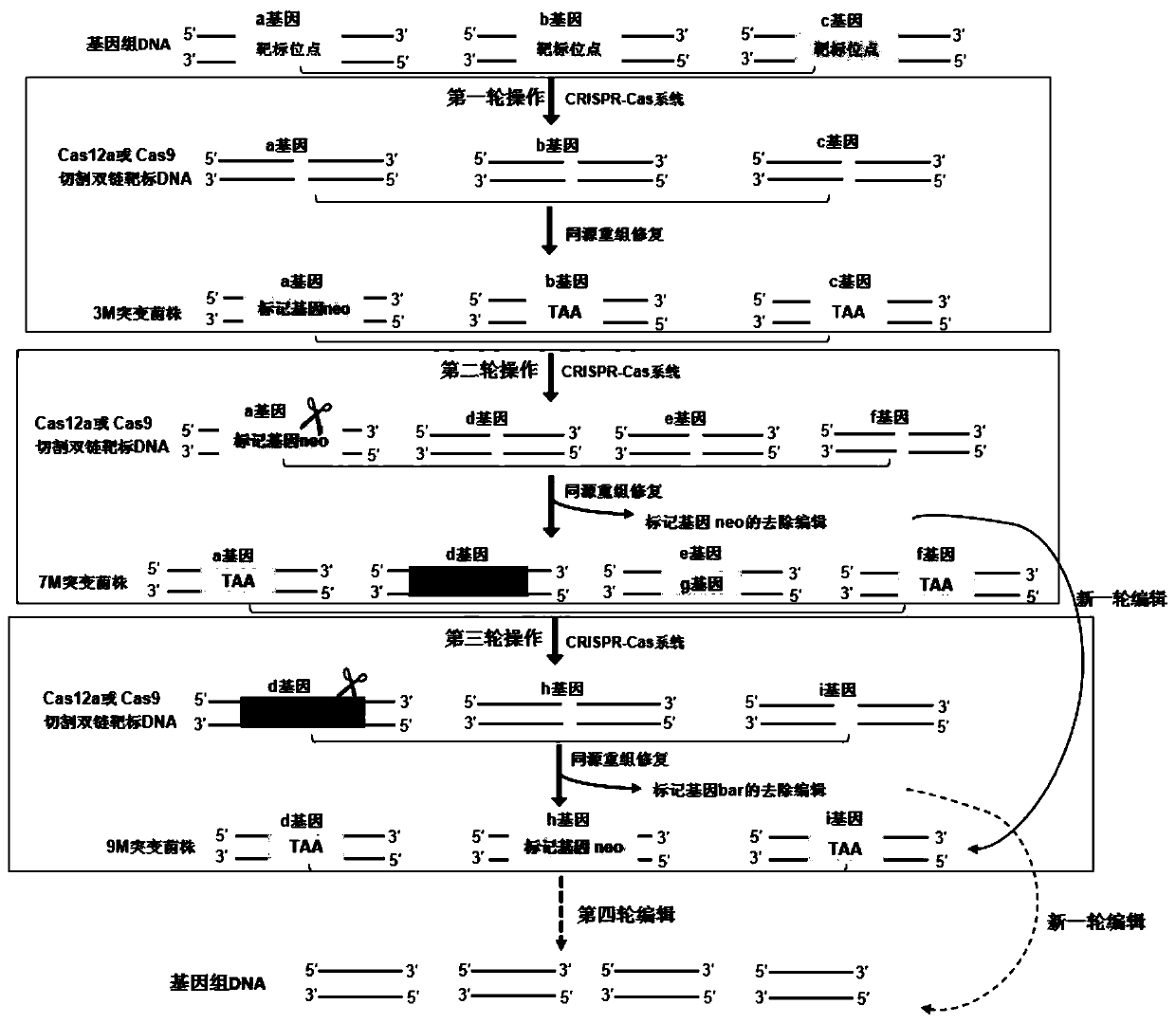
[0097] Example 3, CRISPR-Cas12a (AsCpf1) system multiple rounds of genome editing of Myceliophthora thermophila
[0098] 1. The crRNA tandem plasmid system realizes the second round of genome editing
[0099] The second group of editing system is the Cas enzyme expression cassette Ptef1-AsCpf1-TtprC, the tandem expression element U6p-array2-neo-alp1-rca1-hcr1, and its donor DNA fragments donor-1-alp1, donor-2-gh1-1 , donor-rca1 and donor-hcr1 were mixed at equal molecular molar ratios, and the above three were co-transformed into Myceliophthora thermophila cre1 at a molecular molar ratio of 1:1:1, and the three genes res1 and gh1-1 had been edited at the same time In the first round of mutant strain 3M protoplast cells, Cas12a (AsCpf1) is mediated by crRNA, through protospacer pairing with the DNA strand of the target gene on the host cell genome to recognize the target site for cutting, and then the donor DNA fragment is combined with Homologous recombination occurs between ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com