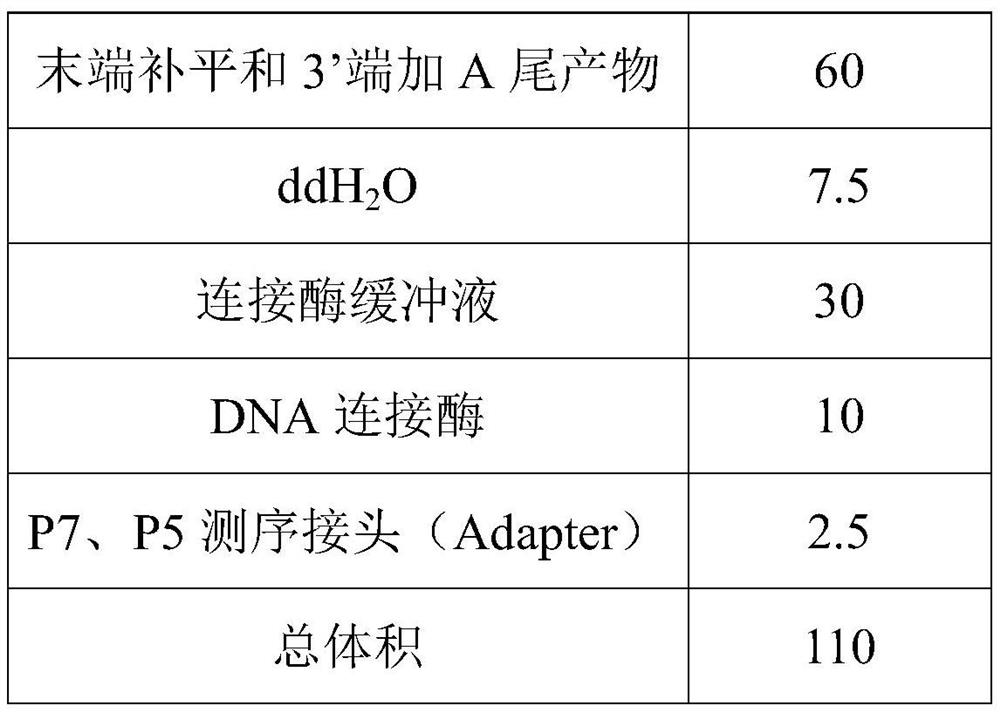
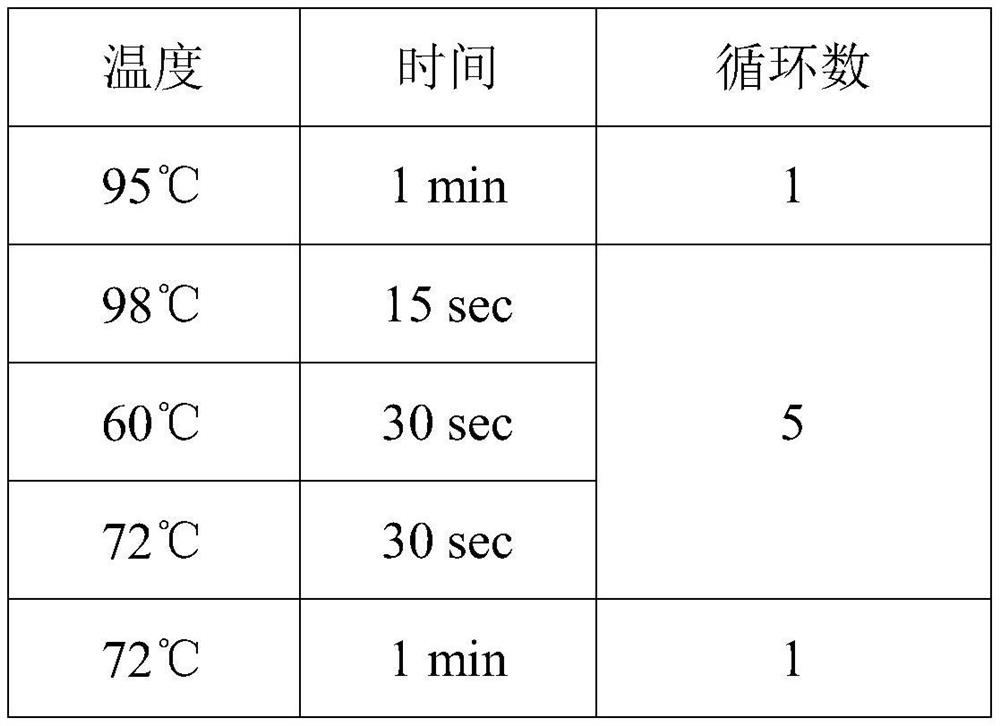
Quantitative fungus detection method based on metagenome sequencing
A metagenomic and genomic technology, applied in the field of quantitative detection of fungi based on metagenomic sequencing, can solve the problems of high proportion of human-sourced reads, large interference of genetic background, and influence on the accuracy of results, so as to reduce the proportion and improve the accuracy of sequencing results interference, low cost, and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1 Sample Pretreatment
[0056] (1) Remove the host genome
[0057] Take 1mL blood sample, centrifuge at 8000g for 3min, collect the precipitate, discard the supernatant, add 200μL PBS to resuspend the precipitate, and vortex to mix;
[0058] Add 200μL 5% saponin and incubate at room temperature for 10min;
[0059] Add 350μL water to incubate for 30s, centrifuge at 6000g for 5min, discard the supernatant;
[0060] Add 100 μL PBS to resuspend the pellet, add 100 μL HL-SAN buffer (5.5M NaCl and 100 mM MgCl 2 );
[0061] Add 5 μL DNaseI and incubate at 37°C for 40 minutes at 1000 rpm;
[0062] Centrifuge at 6000g for 3min, discard the supernatant, and wash twice with 500μL and 1mL PBS respectively;
[0063] Centrifuge at 10,000 g for 1 min, discard 800 μL supernatant, and obtain a sample from which the host genome has been removed;
[0064] (2) Ultrasonic wall breaking treatment
[0065] The sample from which the host genome was removed was ultrasonically bro...
Embodiment 2
[0067] Embodiment 2 sample pretreatment
[0068] (1) Remove the host genome
[0069] Take 1mL of stool sample, centrifuge at 8000g for 3min, collect the precipitate, discard the supernatant, add 200μL PBS to resuspend the precipitate, and vortex to mix;
[0070] Add 200 μL 2% saponin and incubate at room temperature for 15 minutes;
[0071] Add 350μL water to incubate for 30s, centrifuge at 6000g for 5min, discard the supernatant;
[0072] Add 100 μL PBS to resuspend the pellet, add 100 μL HL-SAN buffer (5.5M NaCl and 100 mM MgCl 2 );
[0073] Add 6 μL DNaseI and incubate at 40°C for 30 minutes at 1000 rpm;
[0074] Centrifuge at 6000g for 3min, discard the supernatant, and wash twice with 500μL and 1mL PBS respectively;
[0075] Centrifuge at 10,000 g for 1 min, discard 800 μL supernatant, and obtain a sample from which the host genome has been removed;
[0076] (2) Microwave wall breaking treatment
[0077] The sample from which the host genome was removed was subject...
Embodiment 3
[0079] Example 3 Sample Pretreatment
[0080] (1) Remove the host genome
[0081] Take 1mL soil sample, centrifuge at 8000g for 3min, collect the precipitate, discard the supernatant, add 200μL PBS to resuspend the precipitate, and vortex to mix;
[0082] Add 200 μL 6% saponin and incubate at room temperature for 10 minutes;
[0083] Add 350μL water to incubate for 30s, centrifuge at 6000g for 5min, discard the supernatant;
[0084] Add 100 μL PBS to resuspend the pellet, add 100 μL HL-SAN buffer (5.5M NaCl and 100 mM MgCl 2 );
[0085] Add 2 μL DNaseI and incubate at 35°C for 60 minutes at 1000 rpm;
[0086] Centrifuge at 6000g for 3min, discard the supernatant, and wash twice with 500μL and 1mL PBS respectively;
[0087] Centrifuge at 10,000 g for 1 min, discard 800 μL supernatant, and obtain a sample from which the host genome has been removed;
[0088] (2) Ultrasonic wall breaking treatment
[0089] The sample from which the host genome was removed was subjected to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com