Bacterium expression vector for identifying tumor new antigens and method for screening and identifying tumor new antigens
An expression vector and antigen technology, applied in the field of tumor detection, can solve the problems of non-tumor toxicity and lack of complete specificity of central tolerance, and achieve the effects of high cost, long experimental period and long design time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] This example provides a method for constructing a bacterial expression vector. The method for constructing the carrier specifically includes the following steps:
[0073] (1) clone the hly gene fragment from Listeria species, and clone the hly gene fragment on the ZS91 intermediate expression vector;
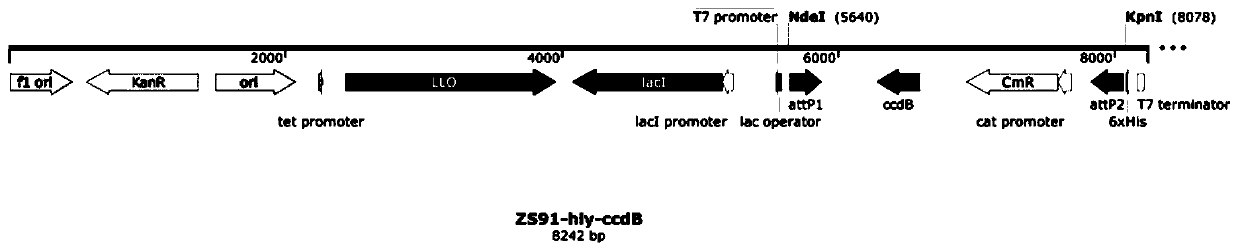
[0074] (2) Insert the ccdB gene fragment on the ZS91 intermediate expression vector in step (1) to increase the efficiency of cloning the neoantigen DNA sequence into ZS91, and refer to the ZS91-hly-ccdB bacterial expression vector diagram after construction figure 1 As shown, the full length is 8242bp.
Embodiment 2
[0076] This example provides a method for testing the bacterial expression vector constructed in Example 1. Specifically include the following steps:
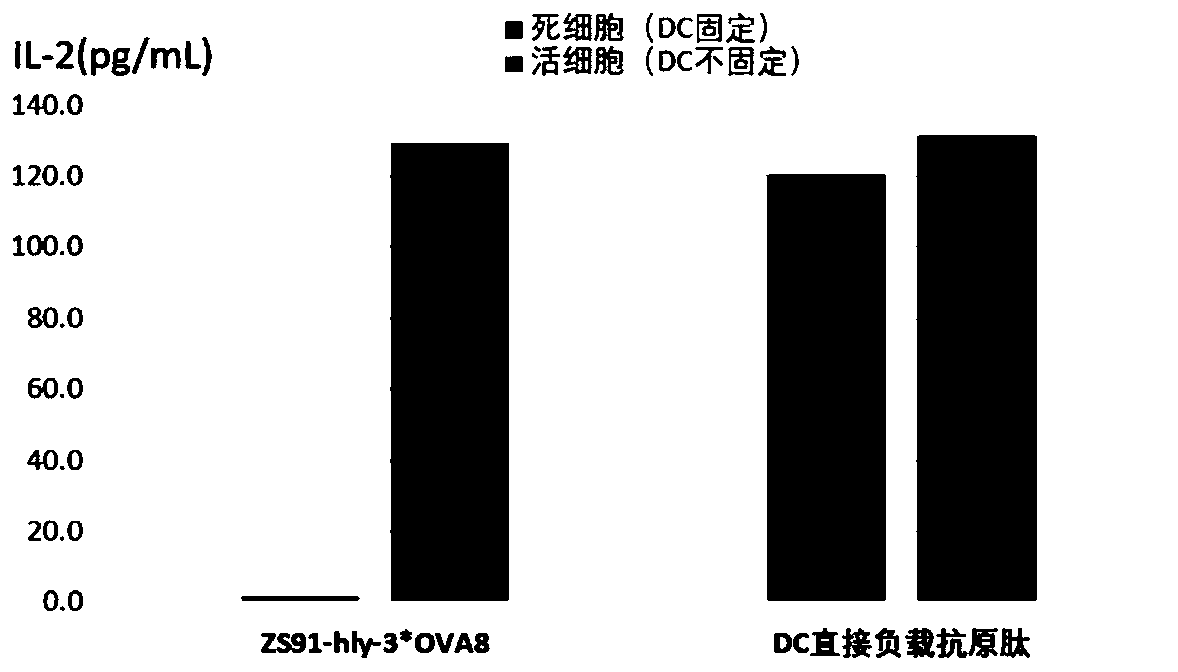
[0077] (1) Synthesis of 3×OVA 8 (EQLE SIINFEKL TEWQLE SIINFEKL TEWQLE SIINFEKL TEW) and CEF sequences. The amino acid sequence of CEF is shown in SEQ ID NO.11, and the DNA sequence of CEF is shown in SEQ ID NO.10.
[0078] (2) Using ELISA to detect the secretion of IL-2 in B3Z cells by 3×OVA8.
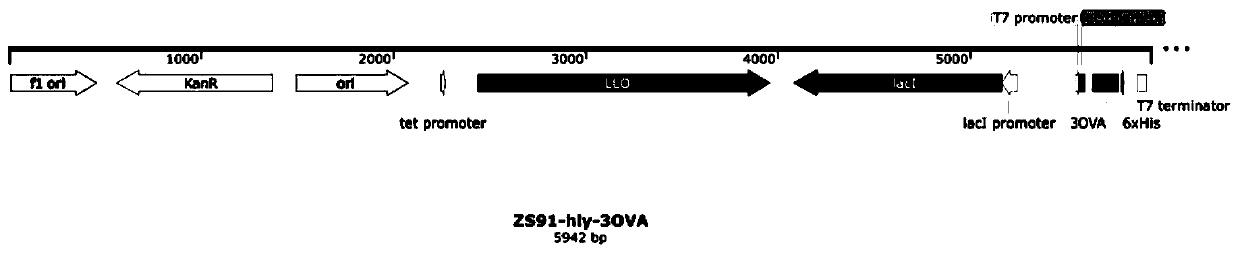
[0079] First, 3×OVA8 was induced to express. On the clone ZS91-hly-3×OVA8 carrying 3×OVA8, the ZS91 empty vector was used as a positive control, and the vector map of the constructed ZS91-hly-3OVA8 was referred to figure 2 As shown, the full length is 5942bp.
[0080] Then transform the constructed ZS91-hly-3OVA8 vector into competent bacteria BL21, pick a single clone and shake the bacteria, dilute the bacterial solution in 100mL LB (Amp) at 1:100, add a final concentration of 1mM IPTG, and induce at 30°C Expression; bacte...
Embodiment 3
[0084] This example provides a method for screening and identifying tumor neoantigens. Specifically include the following steps:
[0085] (1) Discovery and prediction of neoantigens in clinical tumor samples: Tumor tissues were used for whole exome and RNA sequencing to identify mutation sites and expression of mutated genes, using NetMHC, NetCTLpan and IEDB to predict neoantigens, and in vitro according to the prediction results The DNA sequence of the candidate tumor neoantigen was synthesized and cloned into the ZS91 bacterial expression vector constructed in Example 1, and the synthesized DNA sequence of the candidate tumor neoantigen was introduced into the ZS91 bacterial expression vector through Nde I and Kpn I restriction endonucleases.
[0086] (2) Induce expression of candidate tumor neoantigens in bacterial strain BL21.
[0087] (3) Using Elispot to detect the secretion of INF-γ in T lymphocytes.
[0088] The 3 × OVA8 construct was transformed into bacterial compe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com