Codominance marker primer tightly linked with tomato spotted wilt virus resisting sites RTSW of nicotiana alata, identifying method and application of codominance marker primer
A co-dominant marker, spotted wilt technology, applied in the field of molecular biology, can solve the problems of inability to distinguish homozygous/heterozygous genotypes of resistance loci, false positives of SCAR markers, and large-scale application of restriction markers, etc. The effect of obvious difference in belt type, high detection efficiency and lower detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0033]The present invention will be further described in detail below in conjunction with the examples.
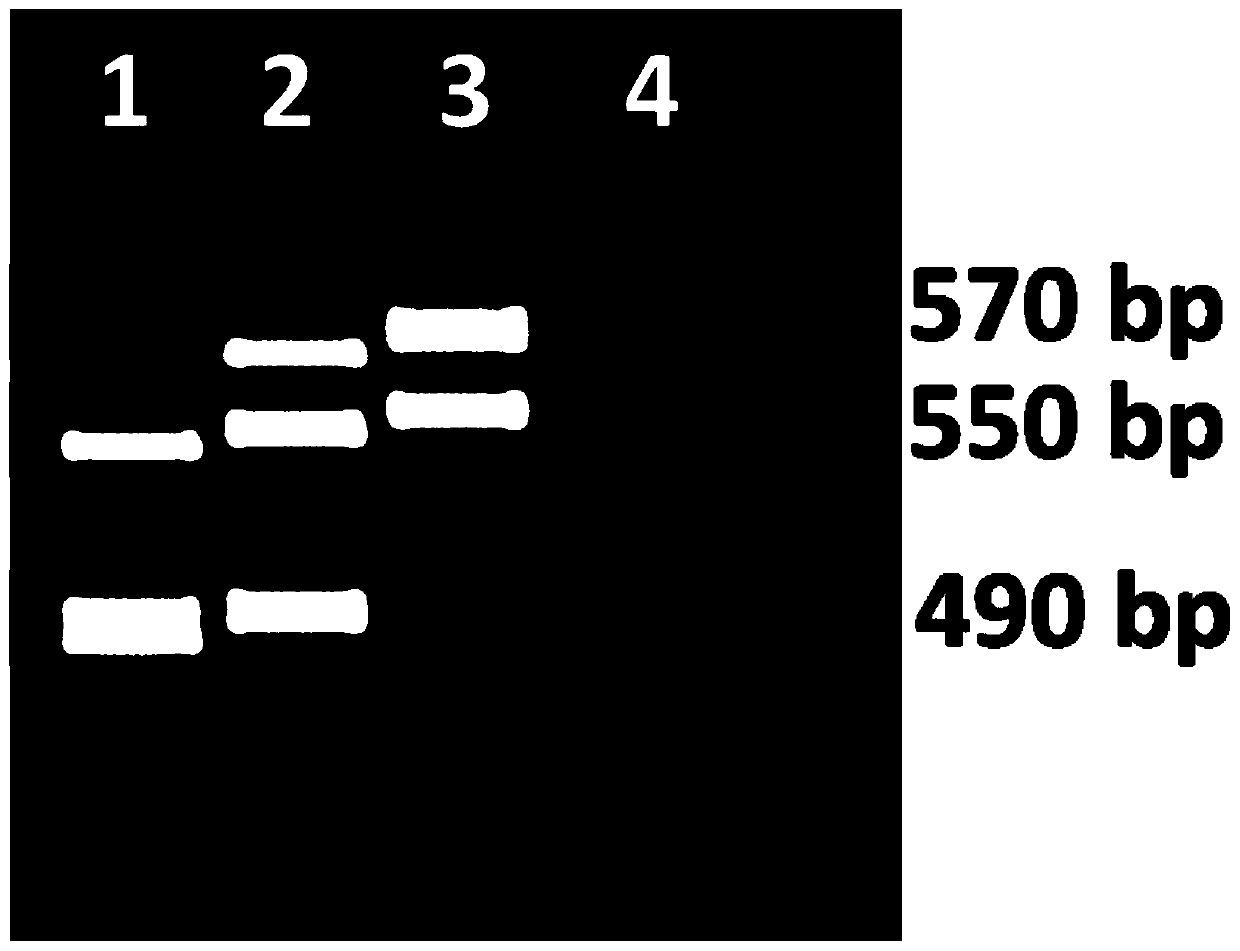
[0034] A co-dominant marker primer closely linked to tobacco spotted wilt locus RTSW, the primer is composed of two single-stranded DNAs of primer 1 and primer 2;
[0035] The primer 1 sequence is Seq ID No.1:
[0036] RTSW_Marker3_F 5'-CCTATGAAGCAACGAAGCGATA-3';
[0037] The primer 2 sequence is Seq ID No.2:
[0038] RTSW_Marker3_R 5'-GTTGACTGTTGACTGTTGATTAGAG-3'.
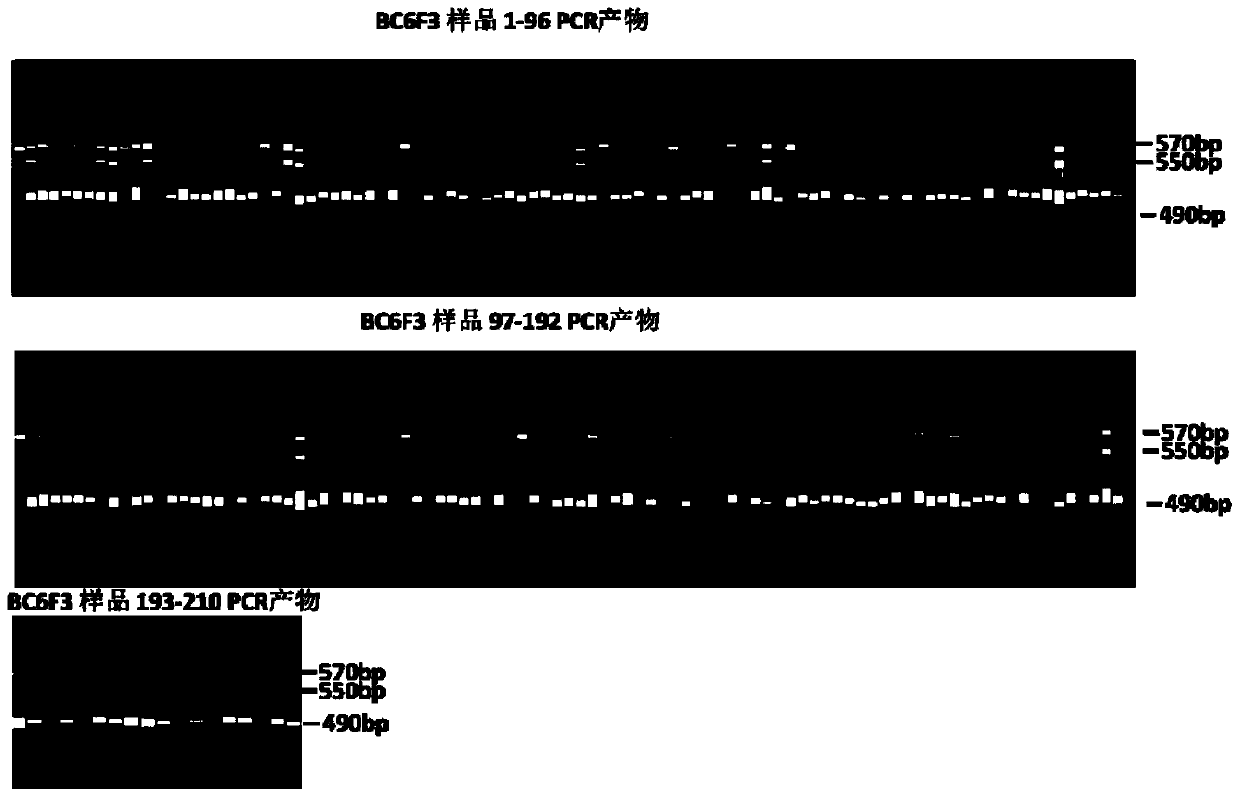
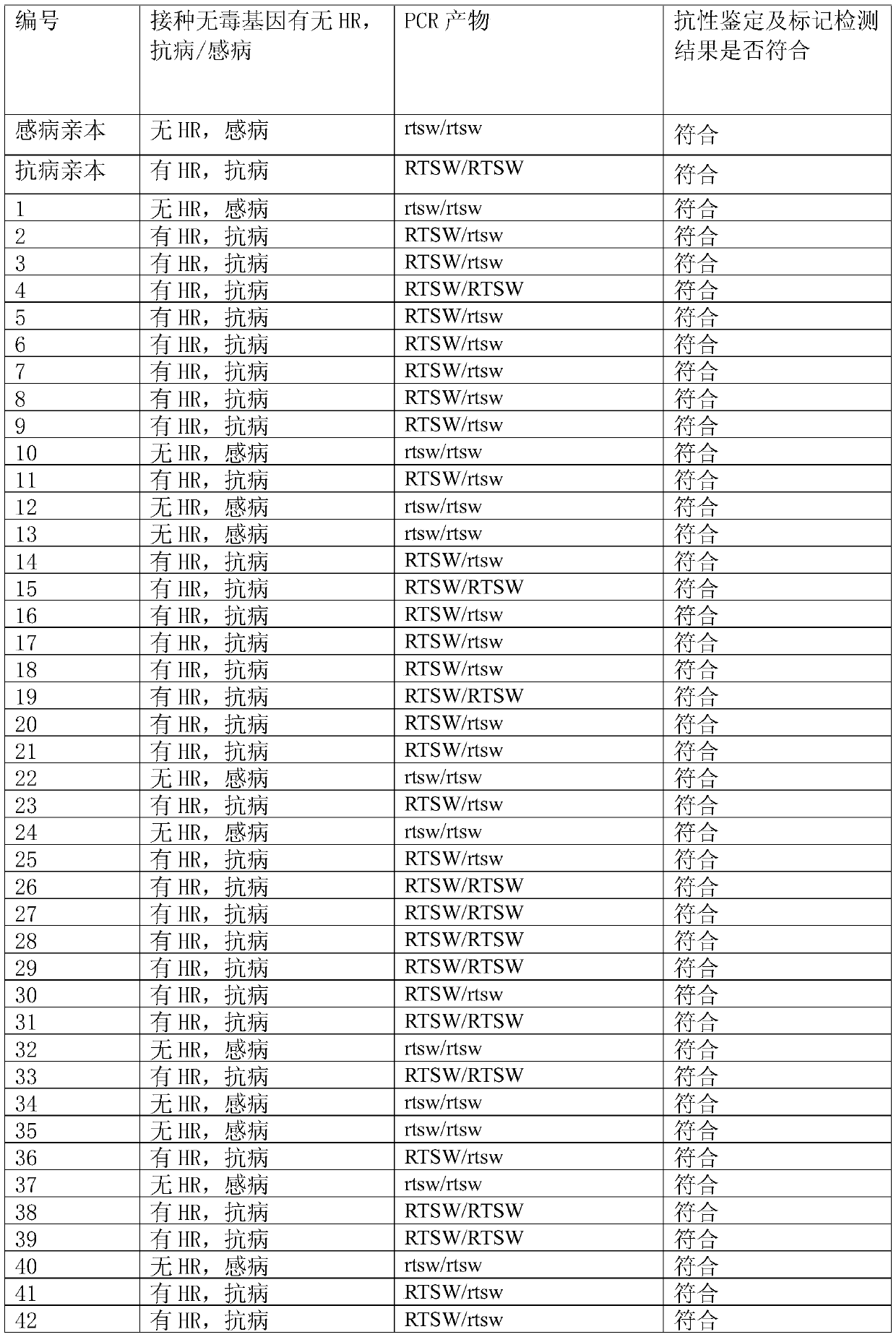
[0039] The method for using the primers to identify the RTSW allele type of the tobacco spot wilt resistance site, the specific steps are as follows:
[0040] (1) Using the genomic DNA of tobacco to be identified, tobacco spotted wilt resistant Polalta and spotted wilt susceptible variety K326 as templates, PCR primers composed of two single-stranded DNAs of primer 1 and primer 2 were used to pair genomic DNA Carry out PCR amplification, and use PCR primers composed of two single-stranded DNAs of primer 1 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com