Tumor mutation analysis method and system, terminal and readable storage medium
An analysis method and tumor technology, applied in sequence analysis, proteomics, instruments, etc., can solve problems such as numerous parameters, complicated process, and insufficient detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
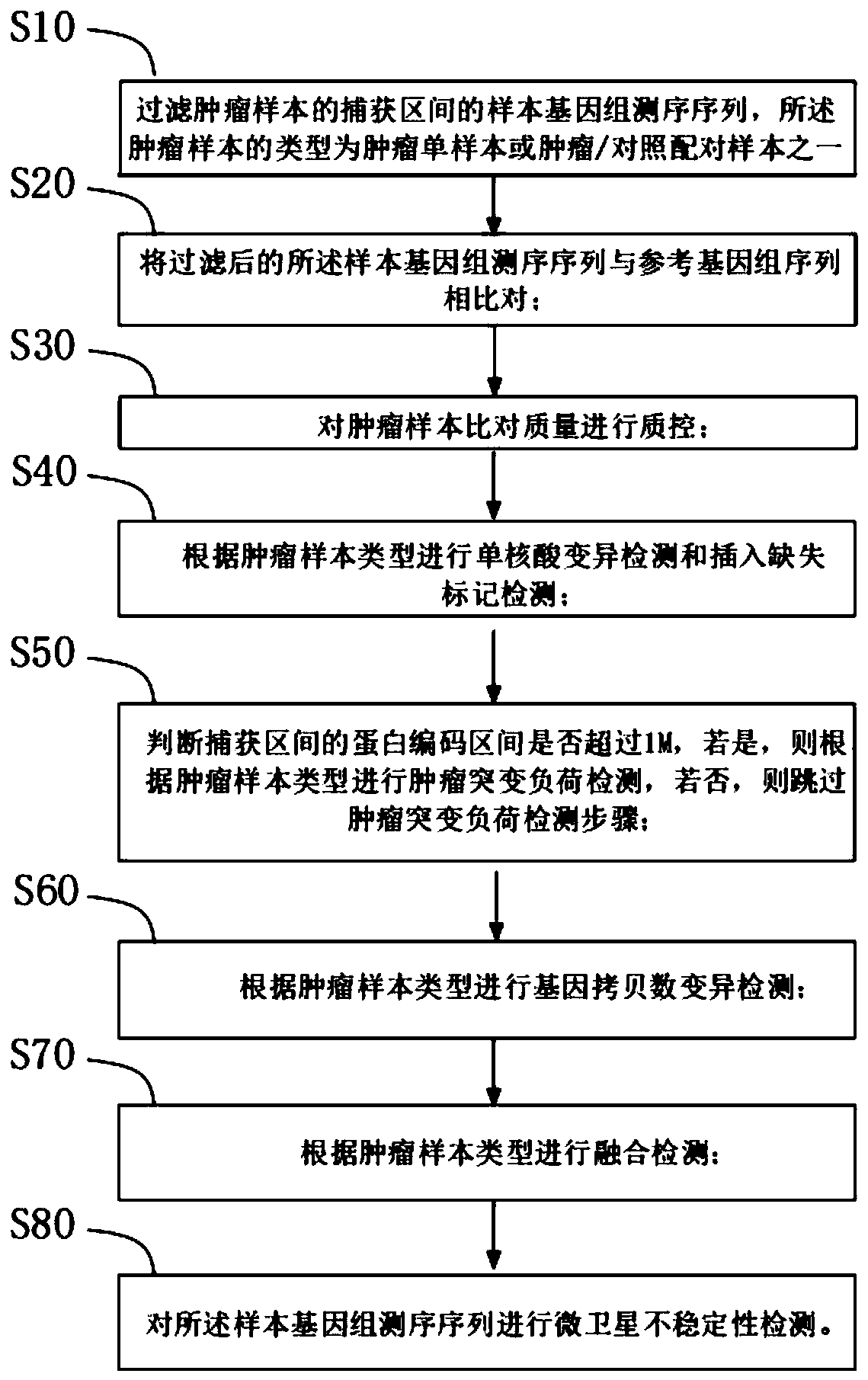
[0052] see figure 1 , a method for analyzing tumor mutations based on next-generation sequencing provided in this embodiment, the method comprising:
[0053] Step S10: filtering the sample genome sequencing sequence in the capture interval of the tumor sample, where the type of the tumor sample is one of a tumor single sample or a tumor / control paired sample;
[0054] Specifically, use fastp to automatically identify and remove the linker sequence contained in the sequence; remove the sequence with poor sequencing quality or high N content, and the specific filtering parameters are -q15-u50-n10. Statistical sequence data volume, Q20, Q30 quality, GC content and other related information.
[0055] Step S20: comparing the filtered sample genome sequence with the reference genome sequence;
[0056] Specifically, ① Align the filtered sequence to the reference genome (hg19) by the BWAmem algorithm
[0057] ② For the result of ①, use MarkDuplicates in picard to remove the repetit...
Embodiment 2
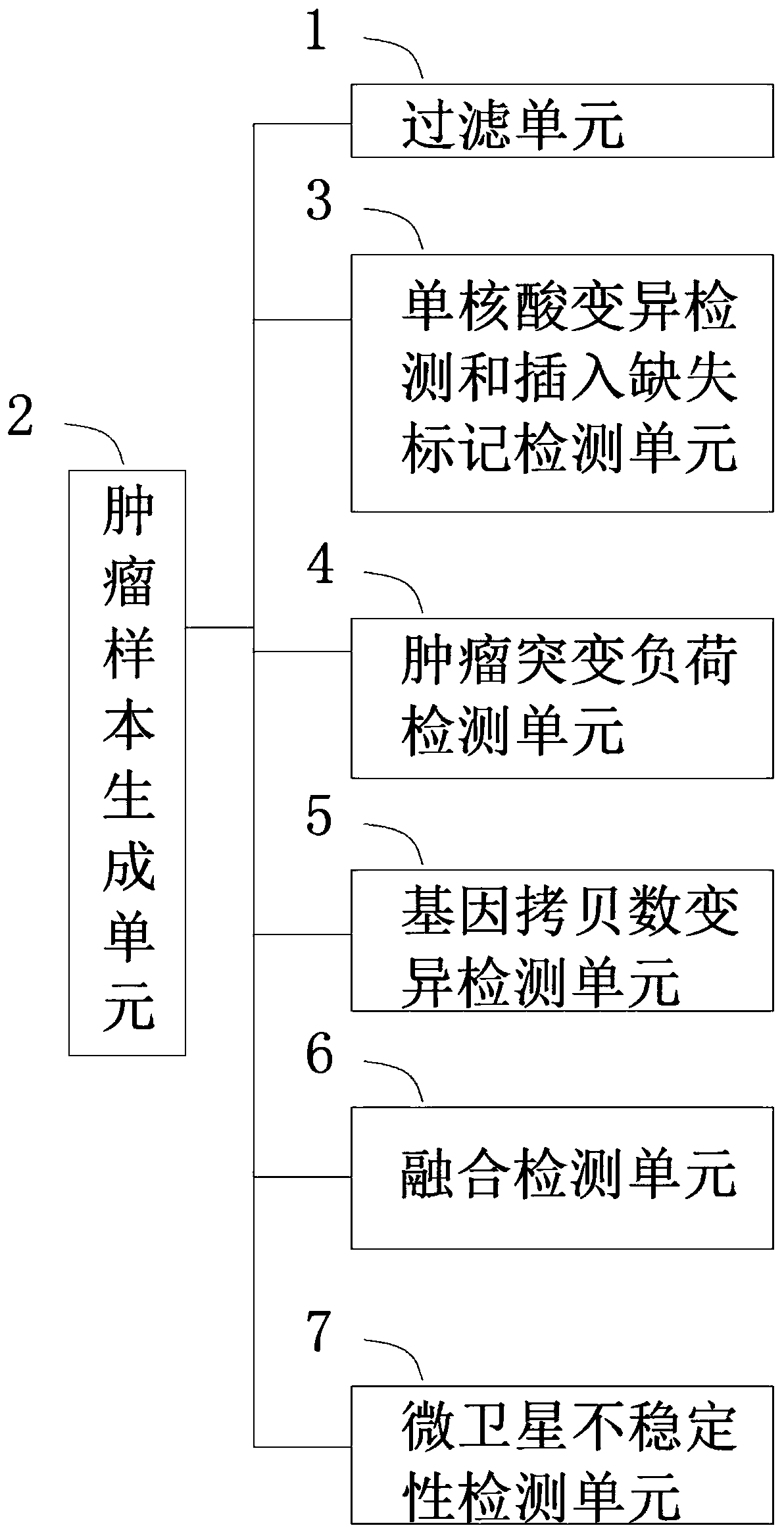
[0118] This embodiment provides a tumor mutation analysis system based on next-generation sequencing, see figure 2 , the system includes:
[0119] A filter unit for filtering the sample genome sequencing sequence;
[0120] The tumor sample comparison and quality control unit compares the filtered sample genome sequencing sequence with the reference genome sequence, and compares the quality control to generate a tumor sample. The type of the tumor sample is a tumor single sample or a tumor / control paired sample one;
[0121] Single nucleic acid variation detection and insertion-deletion marker detection unit, which performs single-nucleic acid variation detection and insertion-deletion marker detection according to tumor sample types;
[0122] The tumor mutation burden detection unit judges whether the protein coding interval of the panel capture interval exceeds 1M, and if so, performs tumor mutation burden detection according to the tumor sample type, and otherwise skips t...
Embodiment 3
[0147] This embodiment provides a control terminal and a computer-readable storage medium applied to the terminal. The computer-readable storage medium stores a computer program, and when the computer program is executed by a processor, the system described in the second embodiment above is implemented. function.
[0148] The terminal comprises a memory, a processor, and a computer program stored in the memory and operable on the processor,
[0149] Exemplarily, a computer program can be divided into one or more modules, and one or more modules are stored in a memory and executed by a processor to implement the present invention. One or more modules may be a series of computer program instruction segments capable of accomplishing specific functions, and the instruction segments are used to describe the execution process of the computer program in the user terminal.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com