Thermostable glucoamylase and methods of use, thereof
A technology of glucoamylase and starch materials, applied in the direction of glycosylation enzymes, biochemical equipment and methods, enzymes, etc., can solve the problems of increasing process costs, thermal stability, slow catalytic activity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0172] Sequence of Penicillium Russell glucoamylase (PruGA1)
[0173] Selection of Penicillium Russell strains available for industrial applications as potential sources of various enzymes. The entire genome of the P. Russell strain was sequenced and the nucleotide sequence of the putative glucoamylase (designated "PruGA1") was determined by sequence identity. The gene encoding PruGA1 is shown in SEQ ID NO: 1:
[0174]
[0175] The amino acid sequence of the PruGA1 precursor protein is shown in SEQ ID NO: 2:
[0176]
[0177] The amino acid sequence of the mature form of PruGA1 confirmed by N-terminal Edman degradation is shown in SEQ ID NO:3:
[0178]
example 2
[0180] Expression and Purification of Penicillium Russell Glucoamylase (PruGA1)
[0181] The nucleotide sequence of the synthetic PruGA1 gene from Penicillium lasselae is shown in SEQ ID NO:4:
[0182]
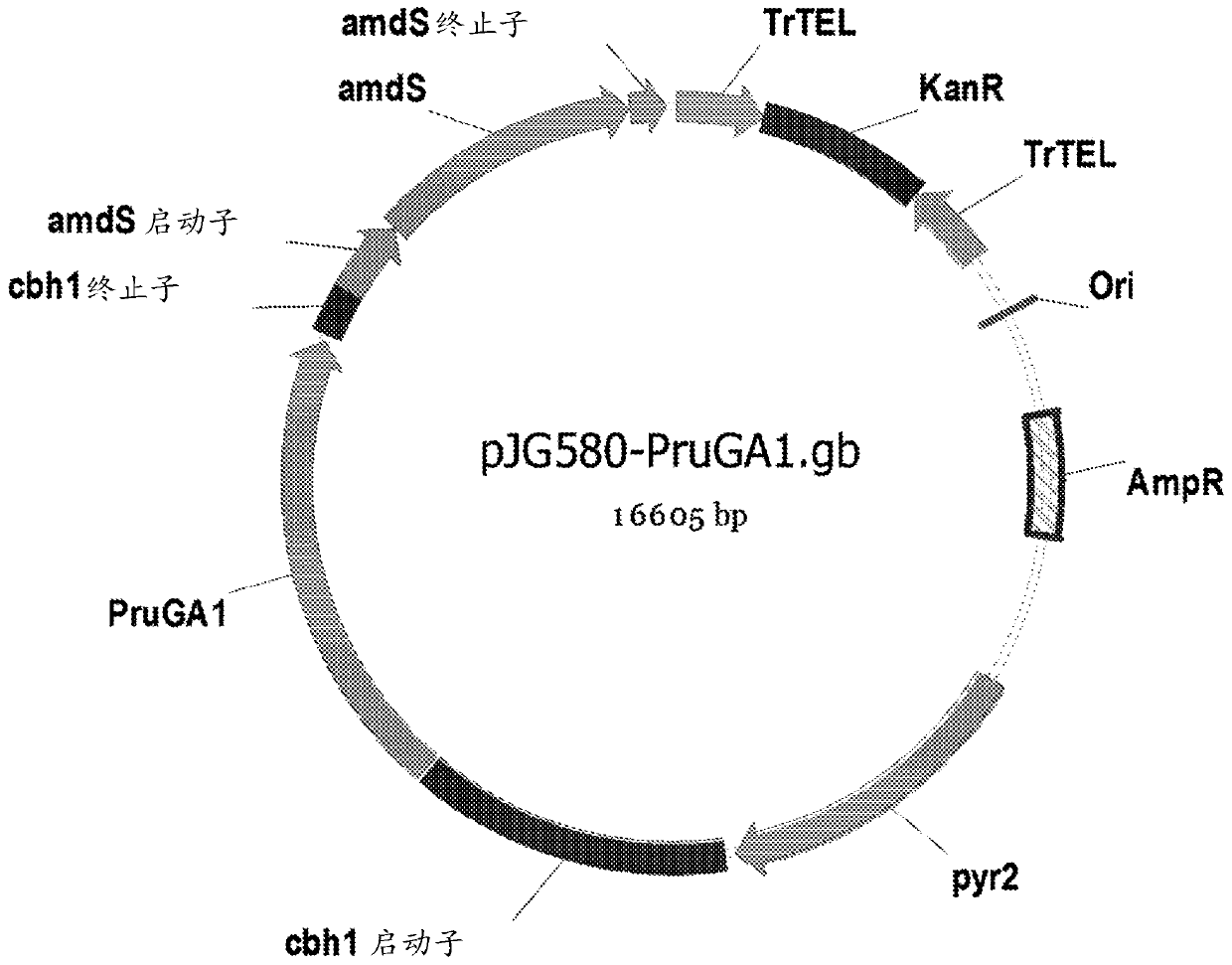
[0183] The DNA sequence of PruGA1 was optimized for expression of PruGA1 in Trichoderma reesei and inserted into the pTrex3gM expression vector (described in US Published Application 2011 / 0136197A1), resulting in pJG580 ( figure 1 ).
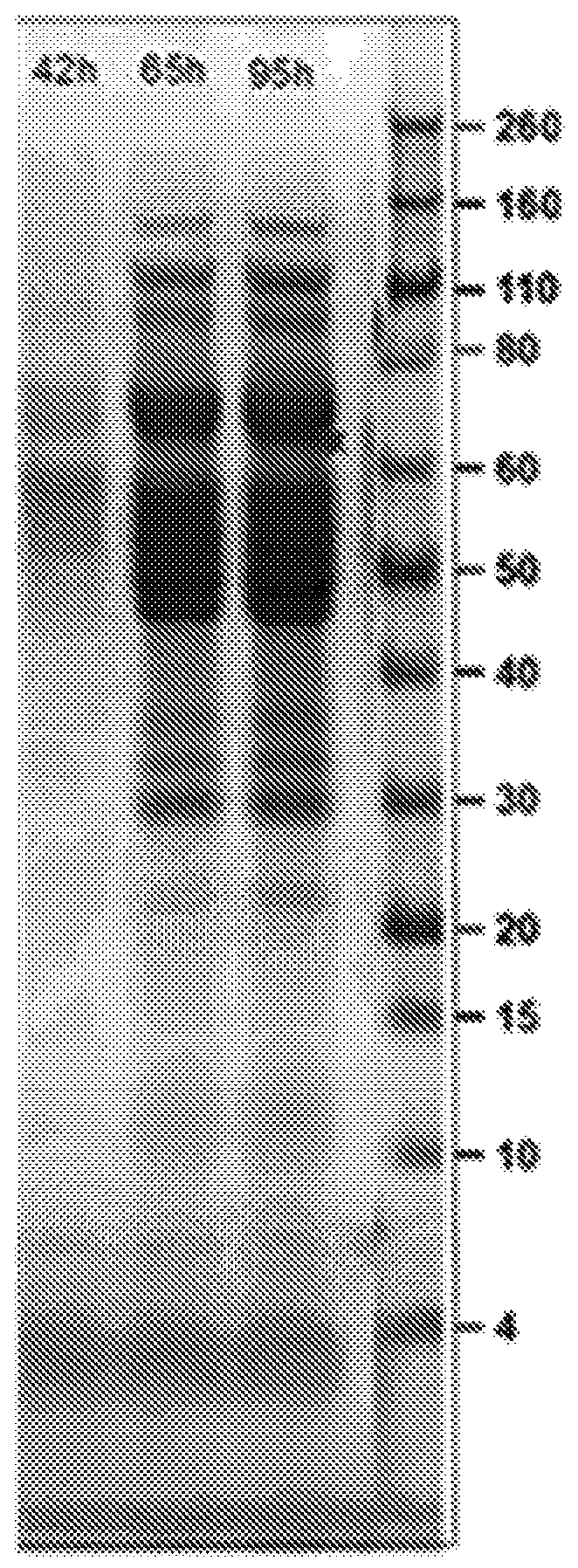
[0184] Plasmid pJG580 was transformed into Trichoderma reesei strain (described in WO05 / 001036) using protoplast transformation (Te'o et al., J. Microbiol. Methods 51:393-99 2002). The transformants described above were selected and fermented by the method described in WO 2016 / 138315. Supernatants from these cultures were used to confirm protein expression by SDS-PAGE analysis and enzyme activity assays.
[0185] Seed cultures of the transformed cells described above were then grown in defined media in 2.8 L fermentors. Fermentation broth...
example 3
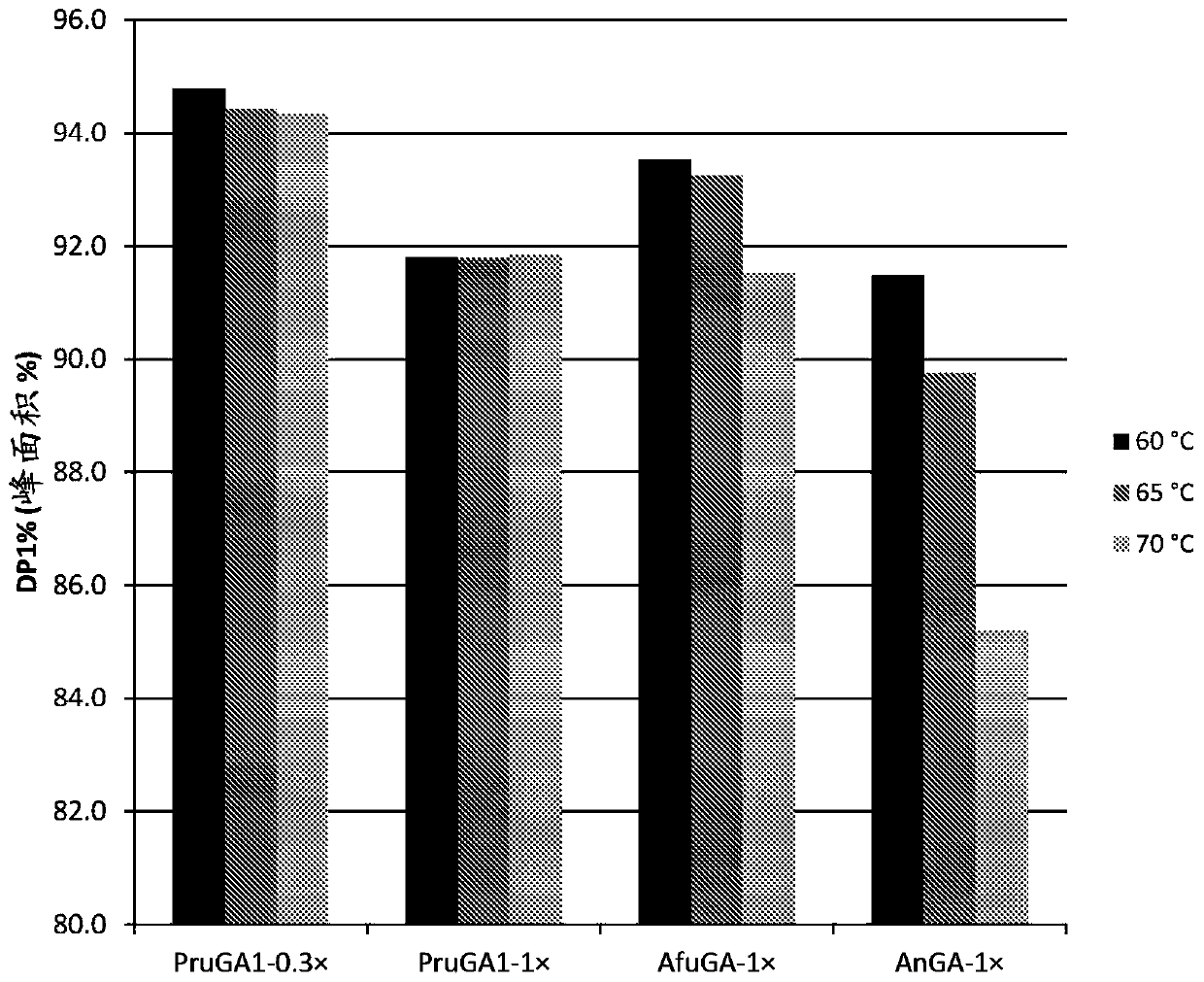
[0188] Specific activity of PruGA1 to soluble starch
[0189] Determination of glucoamylase based on the release of glucose from soluble starch by glucoamylase using the coupled glucose oxidase / peroxidase (GOX / HRP) method (Anal. Biochem. 105 (1980), 389-397) Specific activity.
[0190] Substrate solutions were prepared by mixing 9 mL of soluble starch (1% in water, w / w) and 1 mL of 0.5M sodium acetate buffer, pH 5.0, in a 15-mL conical tube. A solution of coupled enzyme (GOX / HRP) with ABTS was prepared in 50 mM sodium acetate buffer (pH 5.0) with a final concentration of 2.74 mg / mL ABTS, 0.1 U / mL HRP, and 1 U / mL GOX.
[0191] Prepare serial dilutions of glucoamylase samples and glucose standards in purified water. Each glucoamylase sample (10 μL) was transferred to a new microtiter plate (Corning 3641 ) containing 90 μL of substrate solution preincubated at 50° C. at 600 rpm for 5 min. The reaction was carried out at 50 °C for 10 min (shaking (600 rpm) in a thermomixer (Epp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com