Chemiluminescence biosensor for detecting uracil-DNA glycosylase and preparation method and application of chemiluminescence biosensor
A uracil glycosylase and biosensor technology, which is applied in the field of biosensors to achieve the effects of mild reaction conditions, improved efficiency and highly sensitive detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
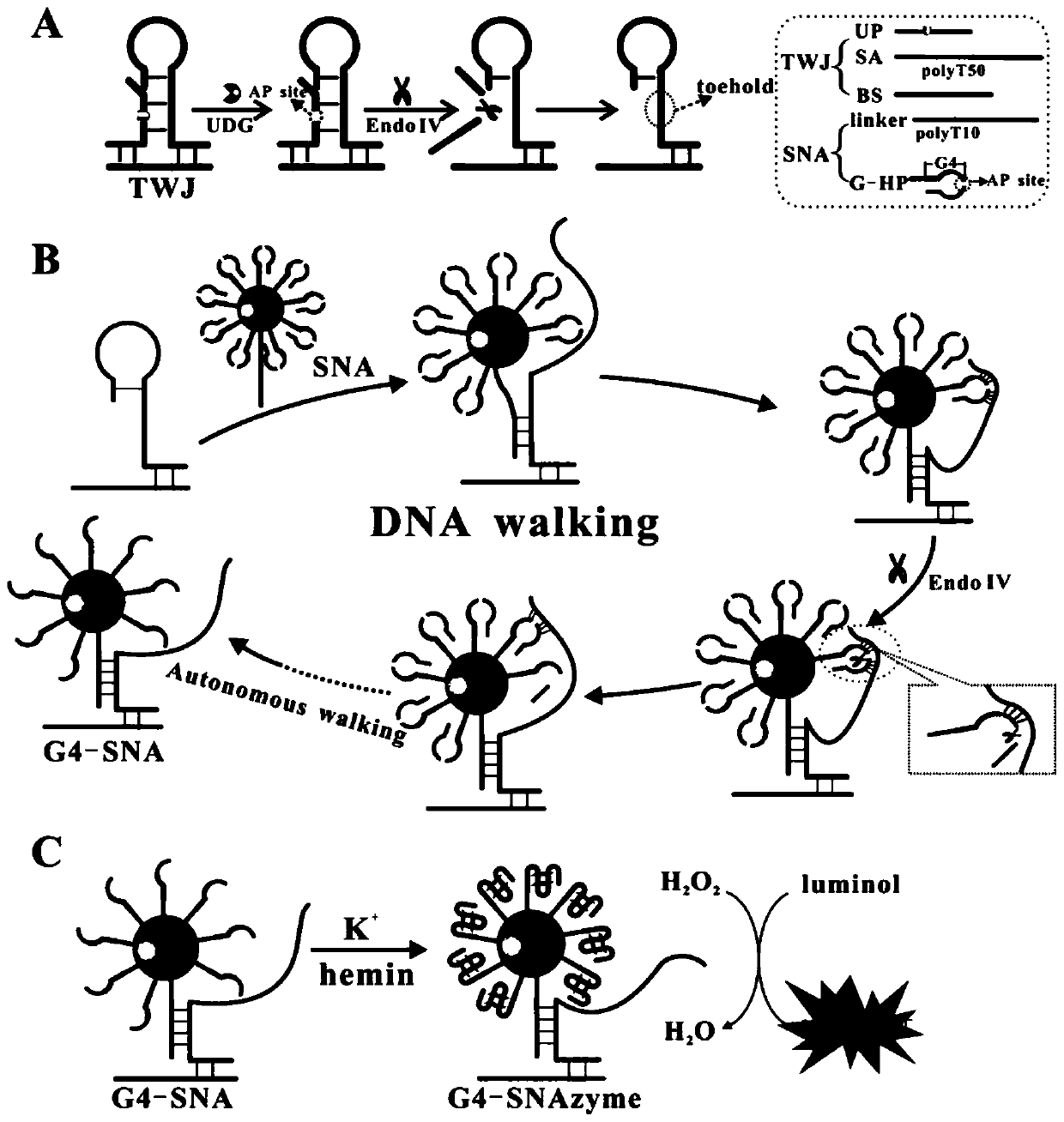
[0051] The preparation method of described biosensor comprises the following steps:
[0052] (1) Preparation of gold nanoparticles;
[0053] (2) Preparation of spherical nucleic acid;
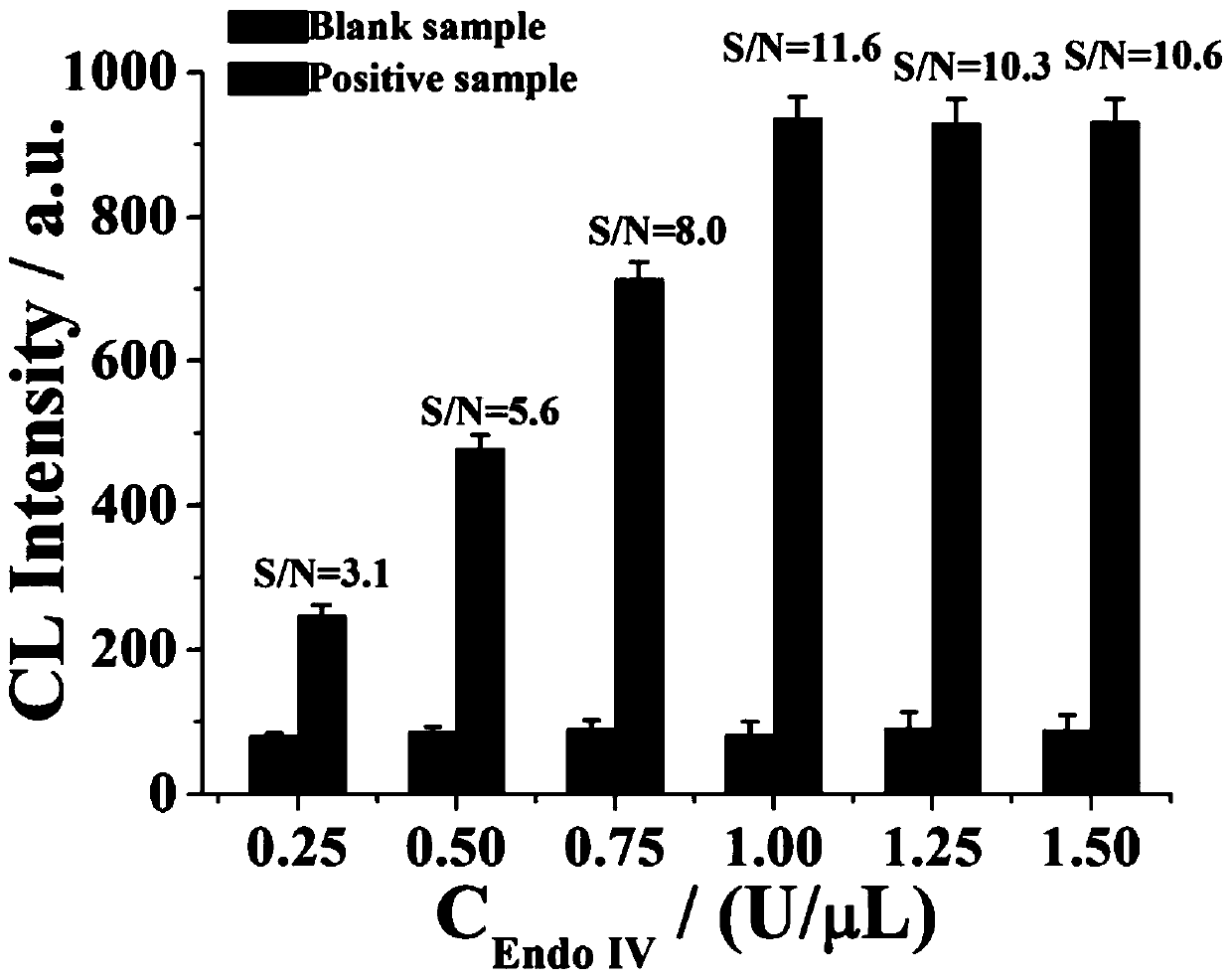
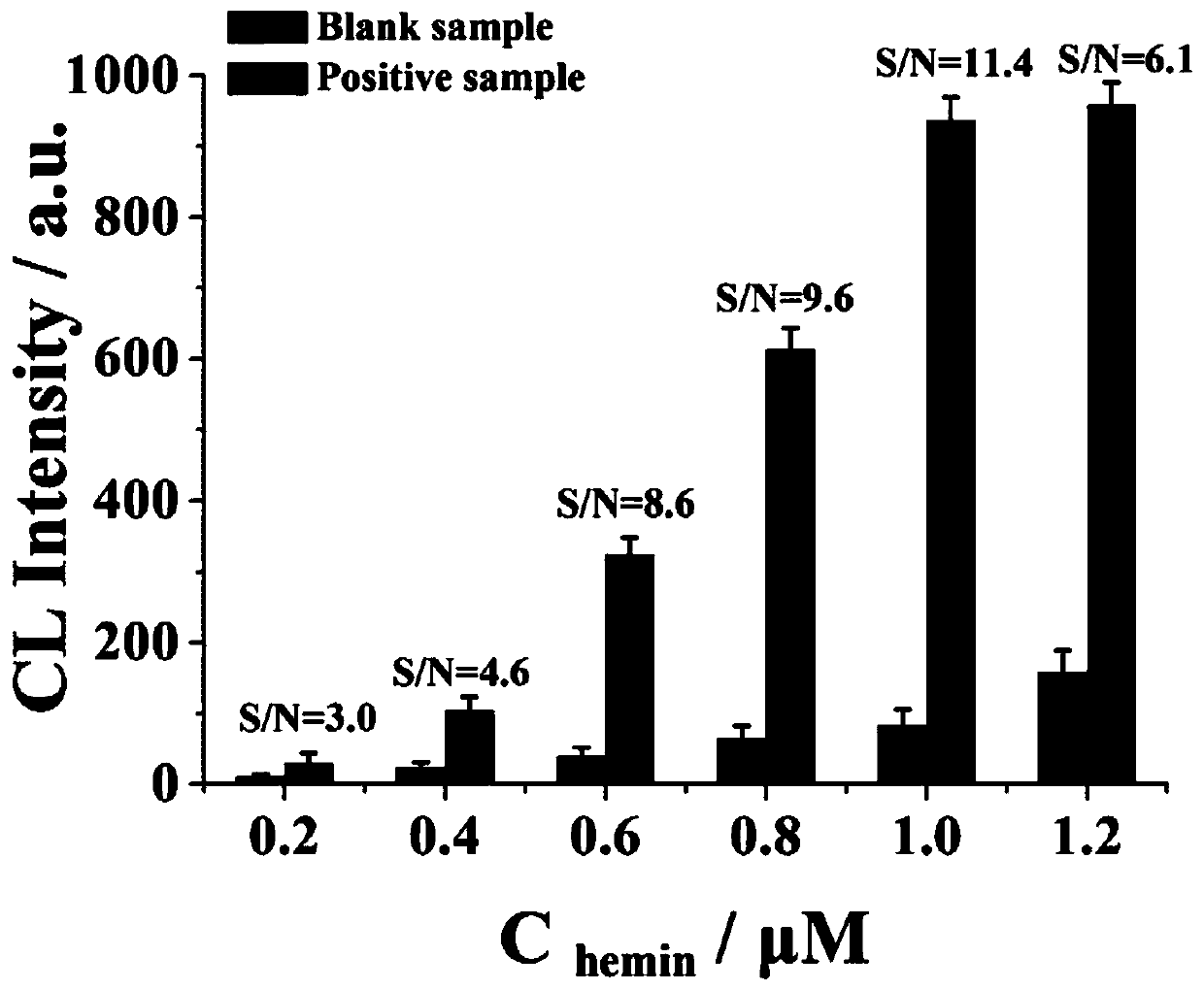
[0054] (3) The formation of spherical nuclease in homogeneous phase is used to catalyze the chemiluminescent reaction of luminol;
[0055] In the described preparation method, the preparation of nano gold:
[0056] The preparation of nano-gold is realized by reducing chloroauric acid with sodium citrate. Add 500μL of chloroauric acid (0.04g / mL) into 200mL of ultrapure water, stir and heat until boiling, then add 3mL of sodium citrate (1%) and quickly add to the boiling solution. Subsequently, it can be observed that the color of the solution changes from light yellow to black and finally to wine red. Continue heating for 15 min after turning wine red to ensure complete reaction. Afterwards, the nano-gold solution was cooled to room temperature and placed at 4°C for use. The UV absorption ...
Embodiment 1
[0058] Preparation of spherical nucleic acids:
[0059] First, the nano-gold stock solution was centrifuged at 13000r / min and 4°C for 20min, then the supernatant was removed and the bottom precipitate was dispersed in ultrapure water to make the concentration 5nM. Then, 150 μL of 10 μM DNA strand (V G-HP:Vlinker =20:1) was added to the nano-gold mixture, and left for 24 hours (4°C) after the addition. Afterwards, 50 μL of PB buffer (10 mM PB, pH 7.4) and 27 μL of PBS (10 mM PB, 2M NaCl, pH 7.4) were added to the mixture. After 48 hours (4°C), continue to add 62 μL of PBS, at this time the concentration of NaCl in the solution is 0.3M. After 24 hours, the mixture was eluted three times by centrifugation at 13000 r / min for 15 minutes to remove unlabeled DNA strands. Finally, the centrifuged precipitate was redissolved in 100 μL of ultrapure water and kept at 4°C for use.
[0060] The spherical nucleic acid has been prepared so far, and the main steps of the reaction process ...
Embodiment 2
[0064] Preparation of spherical nucleic acids:
[0065] First, the nano-gold stock solution was centrifuged at 13000r / min and 4°C for 20min, then the supernatant was removed and the bottom precipitate was dispersed in ultrapure water to make the concentration 5nM. Then, 150 μL of 10 μM DNA strand (V G-HP:Vlinker =20:1) was added to the nano-gold mixture, and left for 24 hours (4°C) after the addition. Afterwards, 50 μL of PB buffer (10 mM PB, pH 7.4) and 27 μL of PBS (10 mM PB, 2M NaCl, pH 7.4) were added to the mixture. After 48 hours (4°C), continue to add 62 μL of PBS, at this time the concentration of NaCl in the solution is 0.3M. After 24 hours, the mixture was centrifuged at 13,000 r / min for 15 min and eluted three times to remove unlabeled DNA strands. Finally, the centrifuged precipitate was redissolved in 100 μL of ultrapure water and kept at 4°C for use.
[0066] The spherical nucleic acid has been prepared so far, and the main steps of the reaction process in th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Extinction coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com