CtDNA library construction and sequencing data analysis methods for simultaneously detecting common mutations of various liver cancers
A construction method and a technology for sequencing libraries, which are applied in DNA preparation, recombinant DNA technology, biochemical equipment and methods, etc., and can solve problems such as time-consuming, inability to detect unknown mutations, and easy contamination.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Embodiment 1, the construction of MC library
[0081] 1. Blunt end repair and A treatment of cfDNA molecules
[0082] Take 10-45ng cfDNA, configure the reaction system as shown in Table 1, and then perform end repair and 3’ end A treatment in the PCR instrument according to the procedures in Table 2 to obtain the reaction product (stored at 4°C).
[0083] Table 1 reaction system
[0084] Element volume cfDNA 50μl End Repair&A-Tailing Buffer(KAPA KK8505) 7μl End Repair&A-Tailing Enzyme Mix(KAPA KK8505) 3μl total capacity 60μl
[0085] Table 2 Reaction program
[0086] temperature time 20℃ 30min 65℃ 30min
[0087] 2. Connection between cfDNA and adapter
[0088] Configure the reaction system according to Table 3, react at 20°C for 15 minutes, and obtain the ligation product (stored at 4°C).
[0089] Table 3 reaction system
[0090] Element volume The reaction product obtained in...
Embodiment 2
[0116] Example 2, RaceSeq enriches the target region and constructs a sequencing library
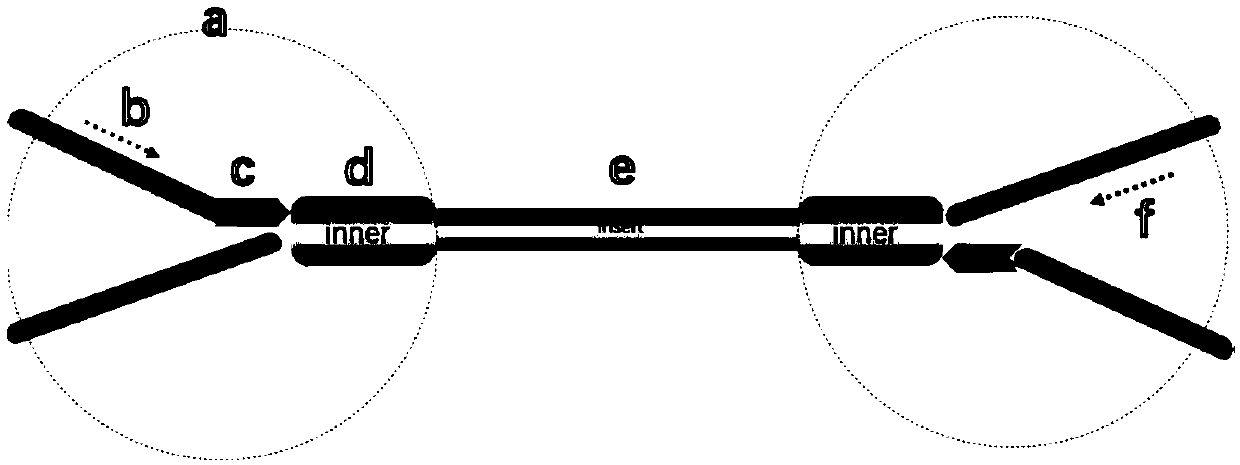
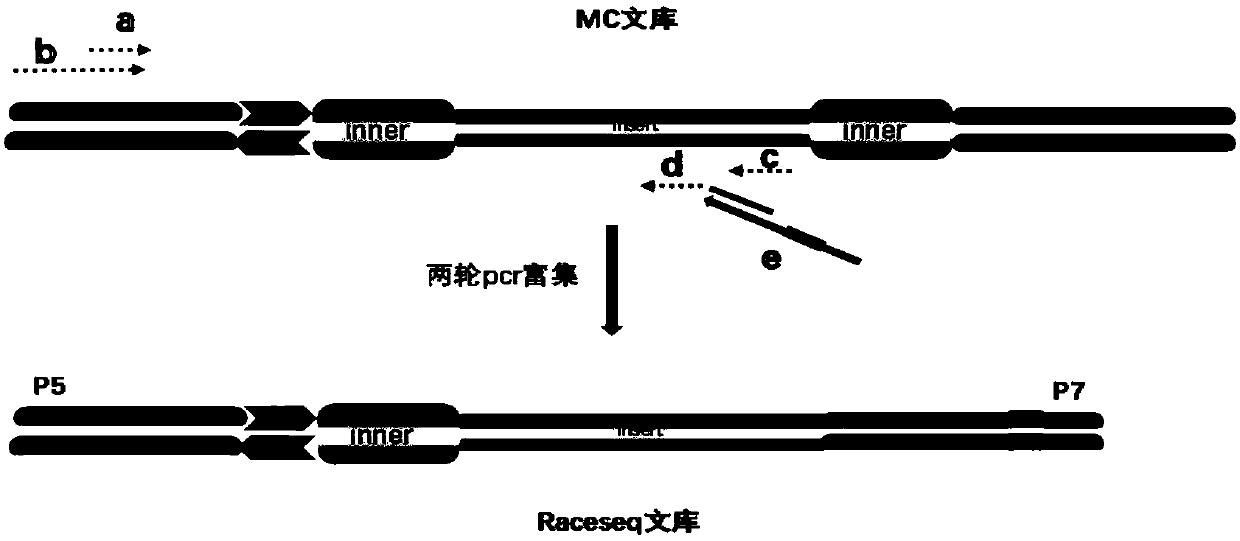
[0117] Such as figure 2 As shown, the MC library was amplified by two rounds of PCR using primers designed for the relevant regions of high-frequency mutation genes of liver cancer in my country (TP53, CTNNB1, AXIN1, TERT) and for HBV integration hotspot regions, and fixed primers. The product is the sequencing library.
[0118] figure 2 Among them, a is the upstream primer of the first round of library amplification, b is the upstream primer of the second round of library amplification, c is the downstream primer library of the first round of library amplification, which is used for the enrichment of specific target sequences, and d is the second round of library amplification Amplify the downstream primer library for the enrichment of specific target sequences, e is the index primer, used to add the index sequence.
[0119] 1. Take 300ng of the MC library prepared in Example 1, div...
Embodiment 3
[0160] Example 3, capture and sequencing of MC library
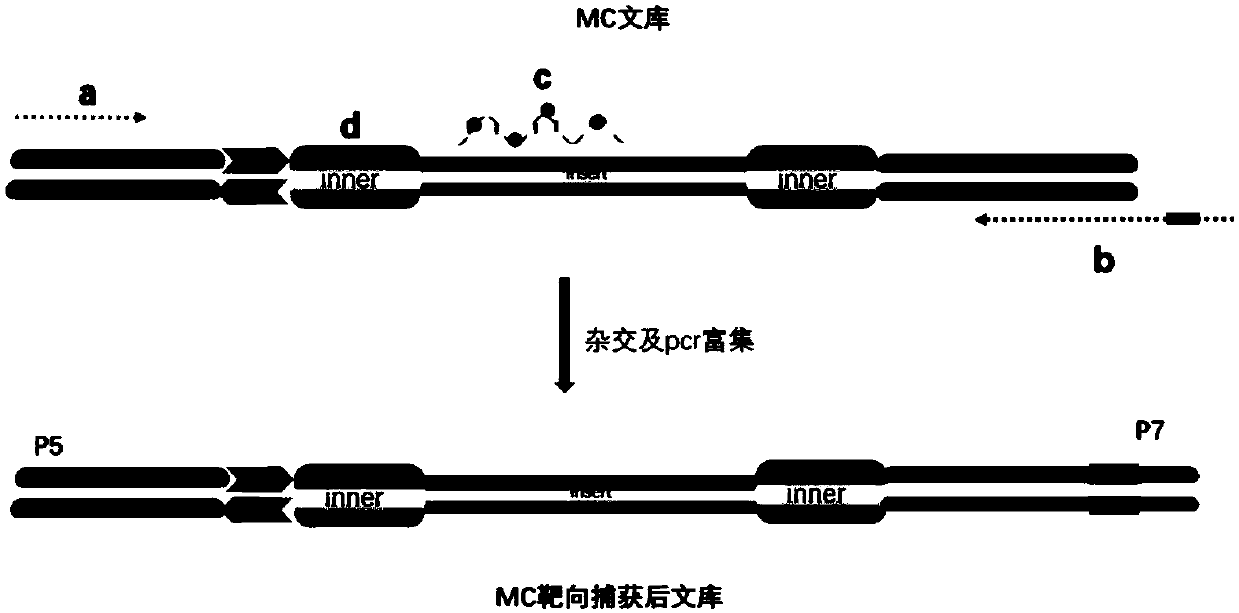
[0161] Such as image 3 As shown, the Agilent sureselect XT targeted capture kit (Agilent5190-8646) can be used to capture the MC library of Example 1 (refer to the kit instructions, and it is also compatible with other brands of capture reagents) to replace the primers in the last step of PCR amplification For the following primers:
[0162] Upstream primer (5'-3'): AATGATACGGCGACCACCGAGATTCTACACTCTTTCCCTACAC GACGCTCTTCC GATCT (Sequence 345)( image 3 In "a"), the underlined part is the same as the primer MC_F, which is used to amplify the library, and the rest is the fixed sequence required for sequencing on the Illumina sequencing platform.
[0163] Downstream primer (5'-3'): CAAGCAGAAGACGGCATACGAGAT (SEQ ID NO: 346)********GTCTC GTGGGCTCGGAGATGTGTATAA (Sequence 347)( image 3 In "b"), the underlined part is the same as the primer MC_R, which is used to amplify the library. ********** is the position of the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com