Crassostrea gigas elongation factor-1-alpha (EF-1-alpha) promoter, and recombinant vector and application thereof
A technology of EF-1 and long oyster, which is applied in the fields of genetic engineering and molecular biology, and can solve problems such as unsuccessful development
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Screening of promoter fragments of housekeeping genes in long oyster.
[0030] 1) According to the published long oyster genome data, six long oyster housekeeping genes, including EF-1α, ACT, 18S rRNA, 28S rRNA and RL7, were preliminarily selected, combined with the existing transcriptome data, to further screen the long oyster embryo stability For highly expressed housekeeping genes, select the EF-1α gene with the highest expression for further analysis;
[0031] 2) Specifically analyze the transcription factor binding site in the front flank sequence of the first ATG of long oyster EF-1α, and finally use the about 3500 bp flank sequence before the ATG including the first exon and the first intron as Candidate promoter regions.
Embodiment 2
[0033] Amplification of the long oyster EF-1α promoter.
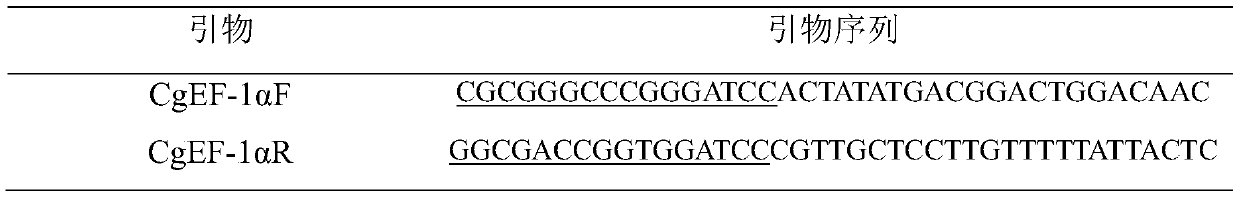
[0034] 1) Genomic DNA of oyster oyster was extracted by phenol-chloroform method, specific primers were designed according to the sequence of the promoter region of the candidate EF-1α gene, and 16 bases of homologous ends of the vector pEGFP-1 were added to the upstream and downstream primers, as shown in Table 1 Show.
[0035] Table 1 Primer sequences used to amplify the promoter of Oyster EF-1α gene
[0036]
[0037] The underlined part is the homologous end of the vector pEGFP-1 with 16 bases.
[0038] 2) Using Phusion High-Fidelity DNA Polymerase from Thermo Scientific Company to perform PCR amplification using the long oyster genomic DNA as a template. The PCR system is shown in Table 2. The PCR reaction program was: pre-denaturation at 98°C for 30s; denaturation at 98°C for 10s, annealing at 64°C for 30s, extension at 72°C for 1min 20s, 35 cycles; extension at 72°C for 10min.
[0039] Table 2 The PCR react...
Embodiment 3
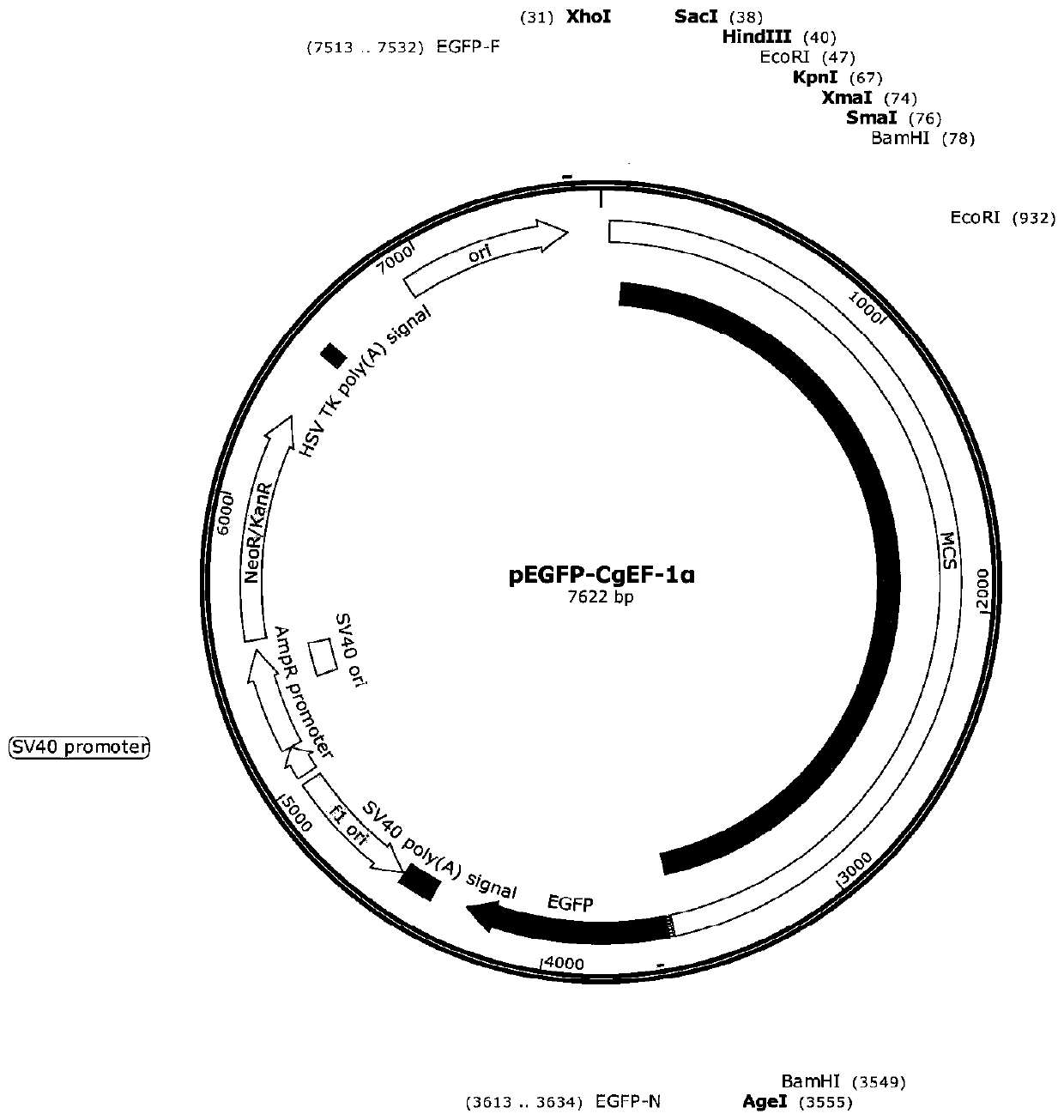
[0043] Construction of pEGFP-CgEF-1α recombinant expression vector.
[0044] 1) The plasmid pEGFP-1 was digested with restriction endonuclease BamHI from New England BioLabs, and the restriction system is shown in Table 3. The reaction program is 37°C, 2h.
[0045] Table 3 Reaction system of BamHI digestion plasmid pEGFP-1
[0046]
[0047]2) The linearized pEGFP-1 was recovered by gel cutting, and In-Fusion ligation (In-Fusion HD Cloning Kit, TaKaRa) was performed with the above PCR recovery product. The In-Fusion connection system is shown in Table 4, and the reaction program is 50°C, 15min.
[0048] Table 4 In-Fusion connection system
[0049]
[0050] 3) Take out Escherichia coli DH5α competent cells and store them at -80°C, place them on ice, take 10 μl of the ligation product and add them to 100 μl competent cells, mix well, place on ice for 30 minutes, heat shock at 42°C for 60-90 seconds, and place on ice On 2min. Add 500 μl of SOC medium, and culture at 150...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com