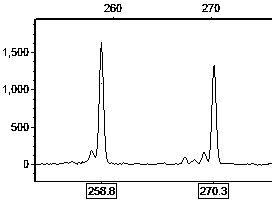
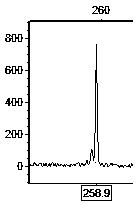
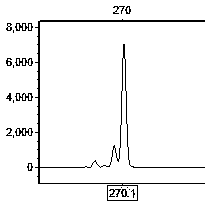
Primer set for hybrid cymbidium tortisepalum EST (expressed sequence tags)-SSR (simple sequence repeats) labeling and screening method
A screening method, the technology of Lily petal orchid, is applied in the primer set and screening field of EST-SSR marker of hybrid Lily petal orchid, it can solve the problems such as not seen, and achieve the effect of convenient statistics, simple operation of the method, and optimized primer ratio
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] The screening method for the primer group marked by hybridization lotus petal orchid EST-SSR, it comprises the steps:
[0046] (1) Perform transcriptome sequencing on the RNA of the target hybrid orchid sample according to the conventional method to obtain the EST tag sequence, and then use MISA software to obtain the sequence segment where all the repeat sequences are located, and establish a repeat sequence database as the original sample of the target hybrid orchid orchid Gene database; the target hybrid lotus petal orchid sample is the blade of an annual plant;
[0047] (2) SSR site search is performed on the original gene database described in step (1), wherein the repeat numbers of one, two, three, four, five, and hexanucleotides are at least 10, 6, 5, 5, and 5 respectively ,5 times;
[0048] (3) Use the upstream and downstream sequences of the SSR site searched in step (2) as the target sequence, use Primer5.0 to design PCR primers for the target sequence, and g...
Embodiment 2
[0058] Embodiment 2: polymorphism analysis of hybrid lotus petal orchid EST-SSR marker
[0059] The polymorphism analysis was performed on the 4 pairs of primer pairs screened by the method described in Example 1 and used for hybridization of the EST-SSR marker of Lianpetalum orchid, and the results are shown in Table 1.
[0060] Table 1 Analysis table of polymorphism of EST-SSR markers in hybrid lotus petal orchid
[0061] Primer pair name Length (bp) total stripe polymorphic band polymorphism information content Primer pair A 248 73 6 0.563 Primer pair B 222 73 3 0.446 Primer pair C 174 75 3 0.4 Primer pair D 274 74 5 0.605
[0062] According to the above structural analysis, 4 pairs of primers can amplify 17 bands, and the number of polymorphic bands is between 3 and 6. On average, each pair of primers amplifies 4.25 polymorphic bands. The morphological ratios are all 100.00%. The polymorphism information content r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com