Primer for simultaneous detection of BRCA 1/2 exon sequence and chemotherapy drug site, and application
An exon and locus technology, applied in DNA/RNA fragmentation, recombinant DNA technology, microbial determination/inspection, etc., can solve the problem of increased risk of breast cancer, high cost of direct sequencing, and large size of BRCA1/2 genes, etc. problems, to achieve the effect of short detection time, low cost, and increased detection range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
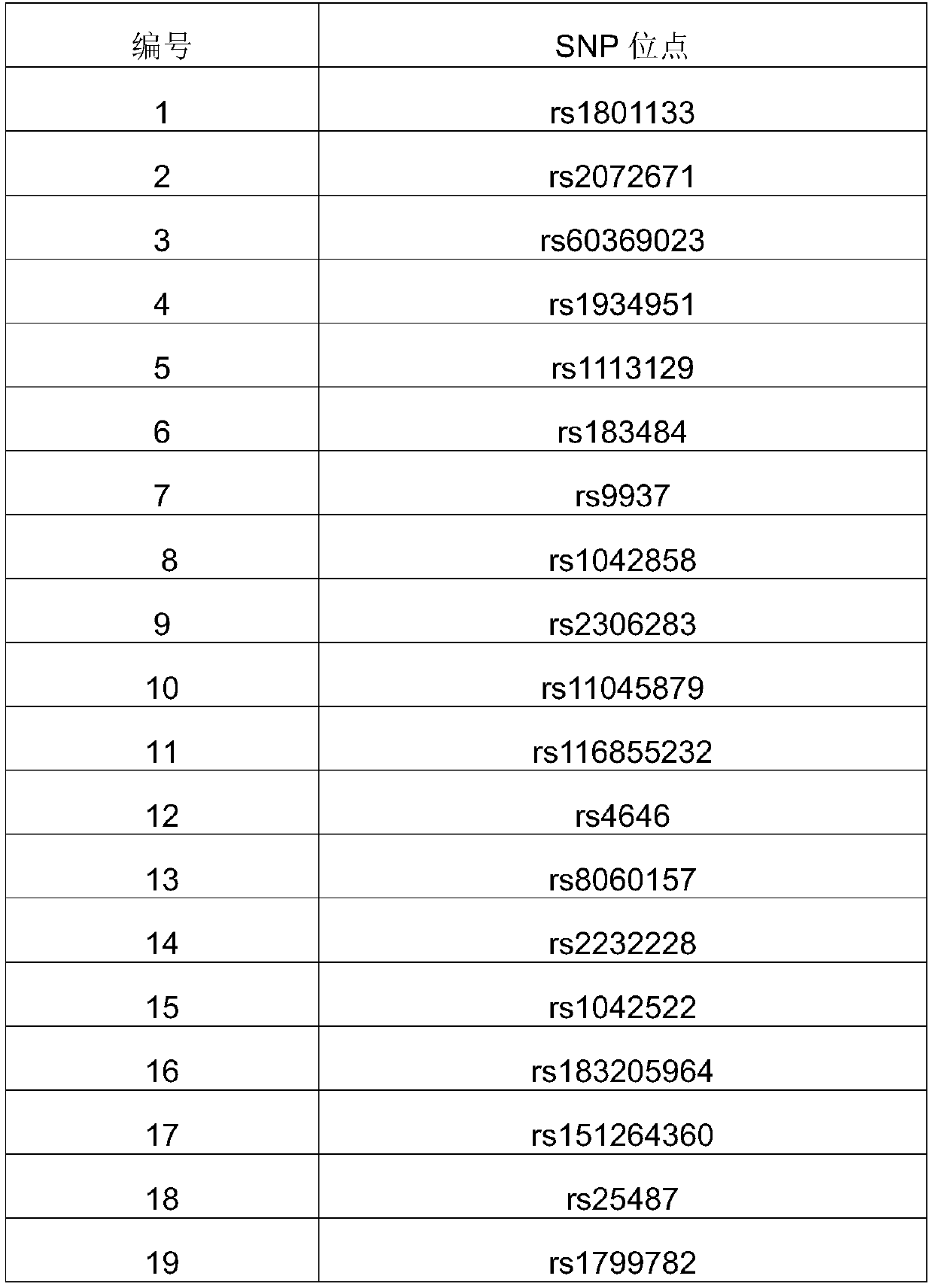
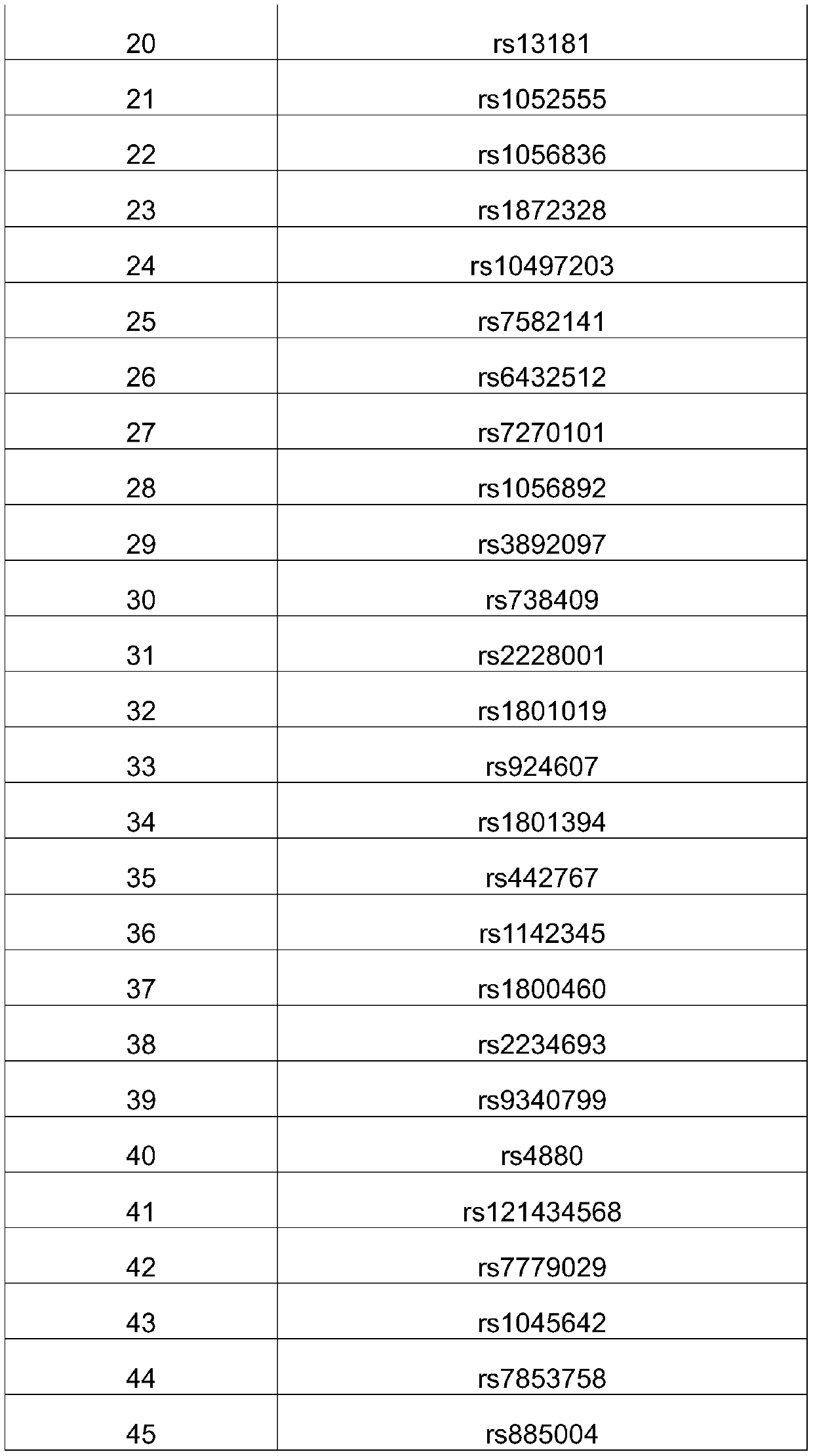
[0040] Use primer3 to design primers with a target amplicon size of 220-300bp to ensure that all primers can cover all exon regions of BRCA1 / 2. Divide the primer pairs of BRCA1 and BRCA2 into two groups to ensure that each set of primers for the same gene There is no overlap of the amplified region inside. Among them, the first and second sets are BRCA1 exon amplification primers: the first set of primers are SEQ ID NO.3-SEQ ID NO.40, and the second set of primers are SEQ ID NO.41-SEQ ID NO. 72. The third and fourth groups are BRCA2 exon amplification primers: the third group of primers are SEQ ID NO.73-SEQ ID NO.130, and the fourth group of primers are SEQ ID NO.131-SEQ ID NO.180.
[0041] Number the 4 primer pairs respectively, number 1, 2, 3...n according to the genome coordinate sequence, and sort them into odd numbered primer pairs with SEQ ID.NO1 added to the 5'end of the upstream primer, and SEQ ID.NO2 added to the 5'end of the downstream primer. , The sequence is that e...
Embodiment 2
[0054] The primers of the present invention perform multiplex PCR amplification methods as follows:
[0055] 1) One round of PCR: use any set of primers from combinations I to IV for PCR reaction, the preferred PCR program is 95°C for 5min; 95°C for 30s, 60°C for 30s, 58°C for 30s, 56°C for 30s, 70°C for 1min (3cycles); 95℃30s, 68℃1min (15cycles); 72℃5min; 4℃∞. As shown in table 2:
[0056] Table 2. The first round of PCR amplification reaction system and amplification reaction conditions
[0057]
[0058]
[0059] 2) One round of purification, the PCR product is purified by Ampure beads for the first time, and then Beadsbuffer and water are used for the second purification; specifically: mix two tubes of PCR products into one tube; add 30μL (Ampurebeads) magnetic beads and mix Let it stand for 5 minutes and then place it on a magnetic stand. After clarification, aspirate the supernatant; wash both sides with 80% ethanol and dry; re-dissolve with 15 μL of distilled water for 3 minu...
Embodiment 3
[0068] A 5mL EDTA blood collection tube was used to collect the blood of a healthy volunteer, and the QIAGEN kit was used to extract blood cell gDNA as a DNA template for multiplex PCR amplification. The multiplex PCR technique in Example 2 was used for detection.
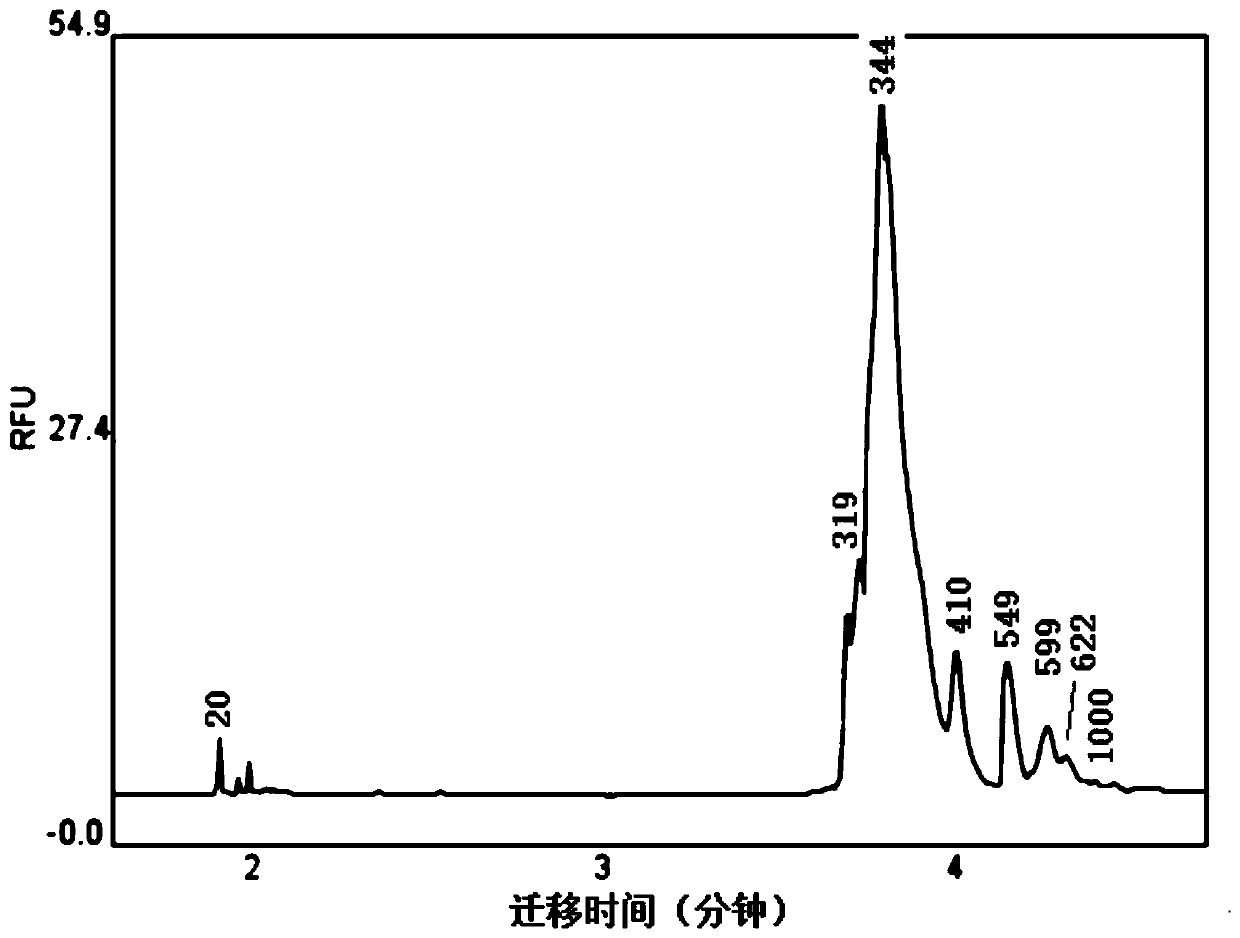
[0069] The concentration of the measured sample is 15ng / μL. Q-Sep results are as figure 1 Shown.
[0070] Table 4, evaluation of offline data quality
[0071] Sequencing raw reads (pieces) Number of reads remaining after quality control (pieces) Proportion of remaining data after quality control 2254879209780293.03%
[0072] The samples were sequenced using the Illumina sequencer. The total number of reads measured was not 2254879. After quality control (removal of unqualified reads), there were 2097802 reads remaining, and the qualified reads accounted for 93.03% of the total reads.
[0073] Table 5, Reference sequence alignment and depth data statistics
[0074]
[0075]
[0076] After the completion of the library ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com