Chondroitinase ac mutant and preparation method thereof
A chondroitin sulfate enzyme and mutant technology, applied in the field of genetic engineering, can solve problems such as low efficiency and limited catalytic activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 Chondroitinase AC mutant protein
[0027] This embodiment provides an AC mutant of chondroitinase, the amino acid sequence of which is shown in SEQ ID NO:2.
[0028] This embodiment also provides a gene encoding the AC mutant of chondroitinase sulfate.
Embodiment 2
[0029] Example 2 Construction of wild-type chondroitinase AC recombinant vector
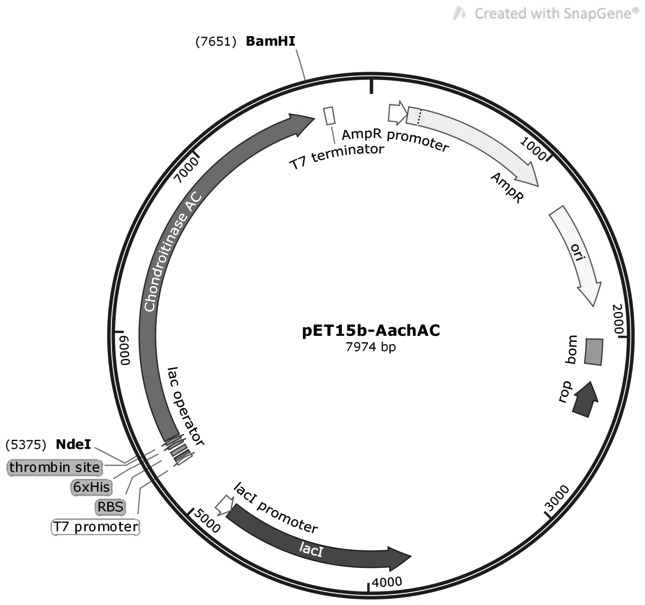
[0030] Using the DNA synthesis sequence corresponding to the amino acid sequence of wild-type chondroitinase AC shown in SEQ ID NO:1 as a template, the original primers were designed for PCR amplification, and NdeI and BamHI restriction enzyme sites were introduced, and then ligated to the plasmid The wild-type chondroitinase AC recombinant vector was constructed by carrying out gene recombination on the vector, and his tag was linked to its N-terminus to facilitate subsequent isolation and purification of the chondroitinase AC mutant. The plasmid vector can be pET15b vector, PMAL-C2X or pGEX-4T1. In this example, the plasmid vector is the pET15b vector, and the wild-type chondroitinase AC recombinant vector is named pET-15b-AachAC. For a schematic diagram, see figure 1 .
[0031] Wherein, the original upstream primer F: 5 ’ -gggaaacatatgGAAGCTGAGCCAGGTGCAGCTG-3 ’ (Among them, the lowercase ...
Embodiment 3
[0033] Example 3 Construction of chondroitinase AC mutant and its recombinant vector
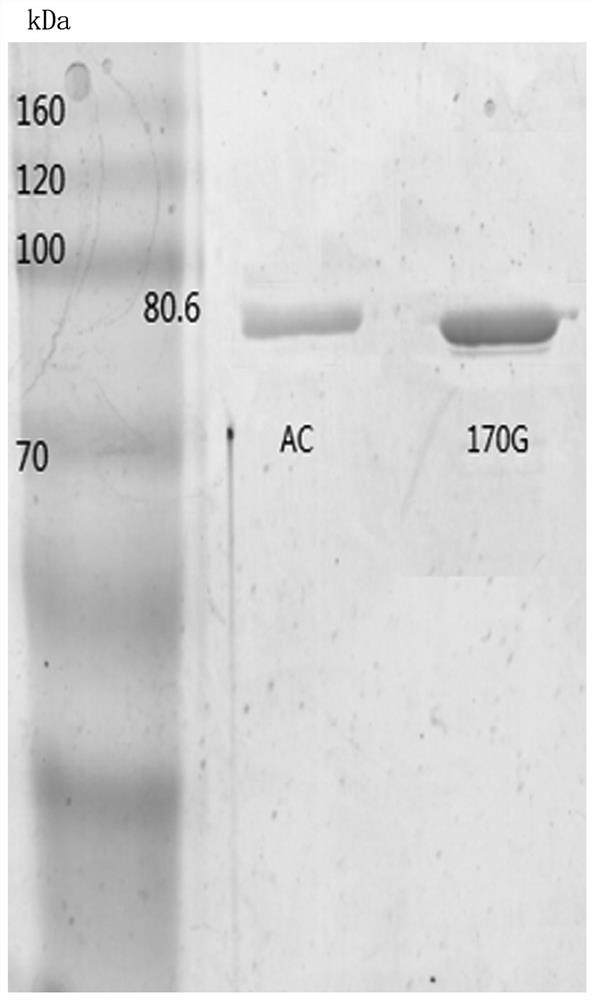
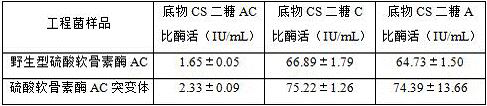
[0034] Using the wild-type chondroitinase AC recombinant vector pET-15b-AachAC provided in Example 2 as a template, the 170th DNA sequence in the amino acid sequence shown in SEQ ID NO: 1 was designed into the upstream primer, Make the 170th position mutated to glycine, and then carry out inverse PCR amplification to obtain the PCR product. The PCR product does not need to be recovered by gel, and is digested with FD DpnI endonuclease at 37°C for 1 h to obtain the chondroitinase AC mutant body The group vector was named pET-15b-AachAC-170G.
[0035] Wherein, the upstream primer F-170G: 5 ’ -GATCCGTGGCTGCAAGGTCCGCCTAAACGT-3 ’ (SEQ ID NO: 3);
[0036] The downstream primer R-170G: 5 ’ -TTGCAGCCACGGATCTGGGACGAAG-3 ’ (SEQ ID NO: 4).
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com