Polypeptide capable of specifically targeting triple-negative breast cancer stem cells and application thereof
A technology for recombining cells and amino acids, applied to medical preparations with non-active ingredients, medical preparations containing active ingredients, peptides, etc., can solve incomplete, lack of specific targets, and hinder triple-negative breast cancer stem cells. For drug development and other issues, achieve high specificity and affinity, and high specificity selection effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] 1. Culture and identification of triple-negative breast cancer stem cells
[0043] A large number of triple-negative breast cancer cells with stem cell characteristics were cultivated by a serum-free low-adsorption suspension culture method, and their stem cell characteristics were verified for subsequent screening of targeted polypeptides.
[0044] The specific operation steps are as follows:
[0045](1) Use serum-free low-adsorption suspension to culture triple-negative breast cancer stem cells. The medium components used include DMEM / F12 medium, epidermal growth factor, basic fibroblast growth factor, insulin, and B27.
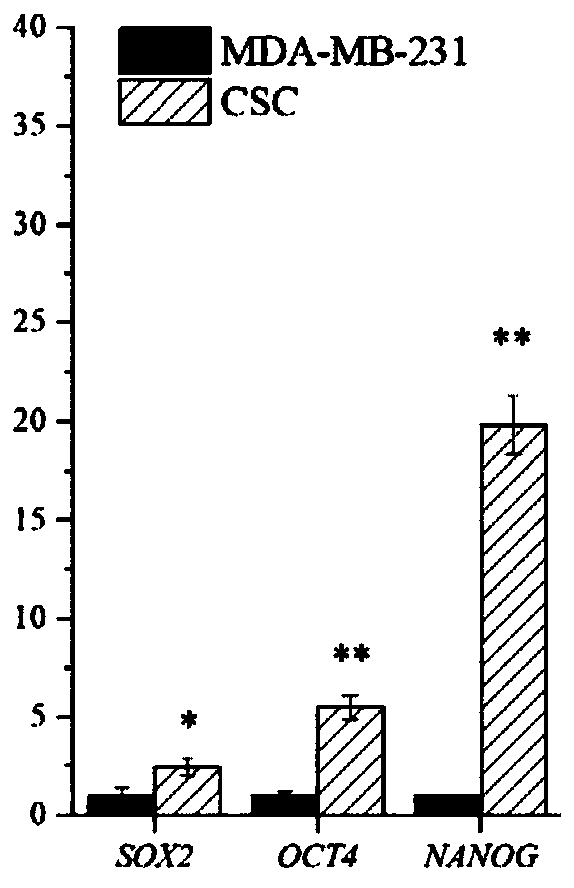
[0046] (2) qRT-PCR verifies the expression levels of biomarkers such as SOX2, OCT4, and NANOG in triple-negative breast cancer stem cells to verify whether they have stem cell characteristics.
[0047] figure 1 It is the result of the relative expression of stem cell-related genes SOX2, OCT4, and NANOG in triple-negative breast cancer stem cells (...
Embodiment 2
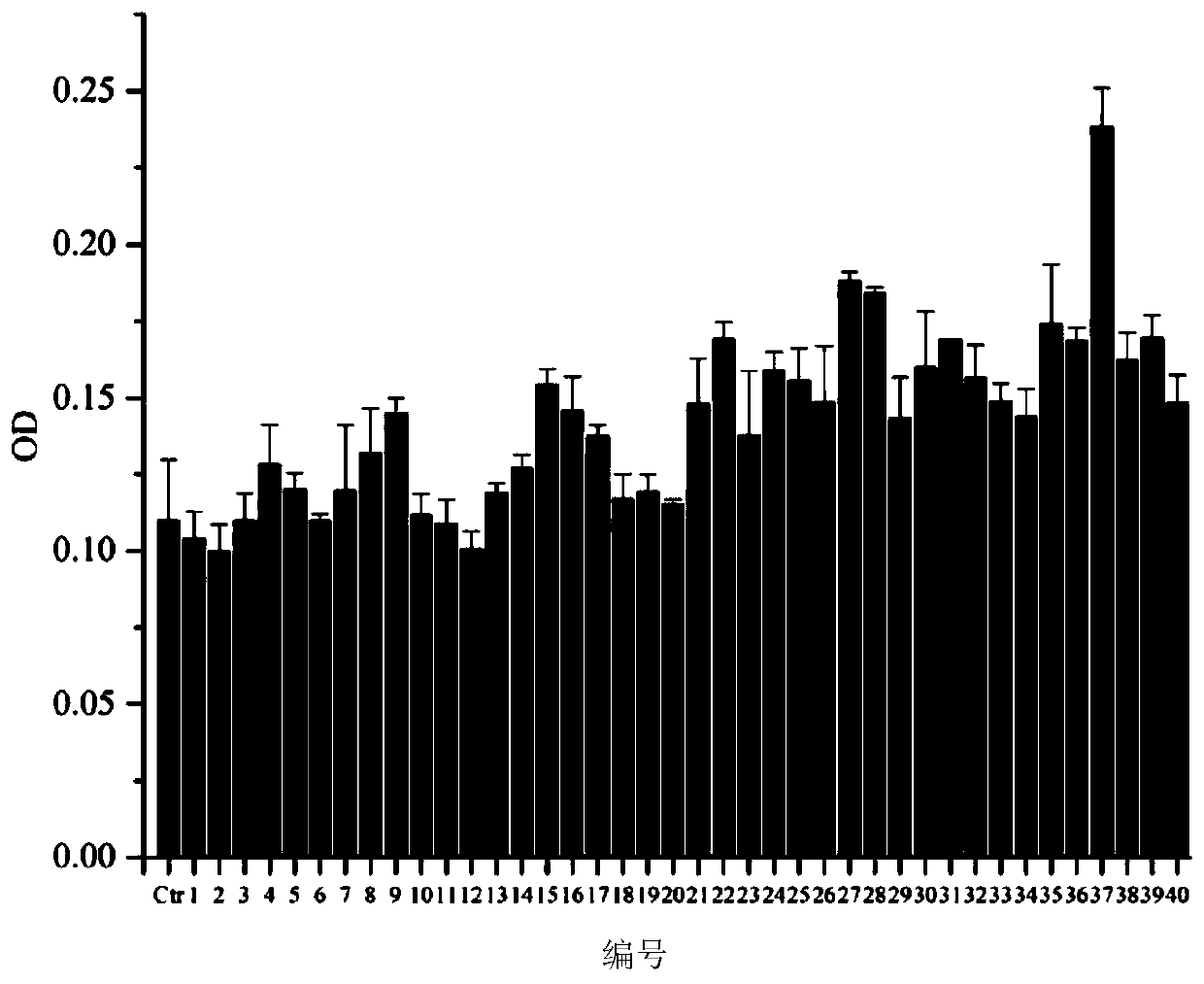
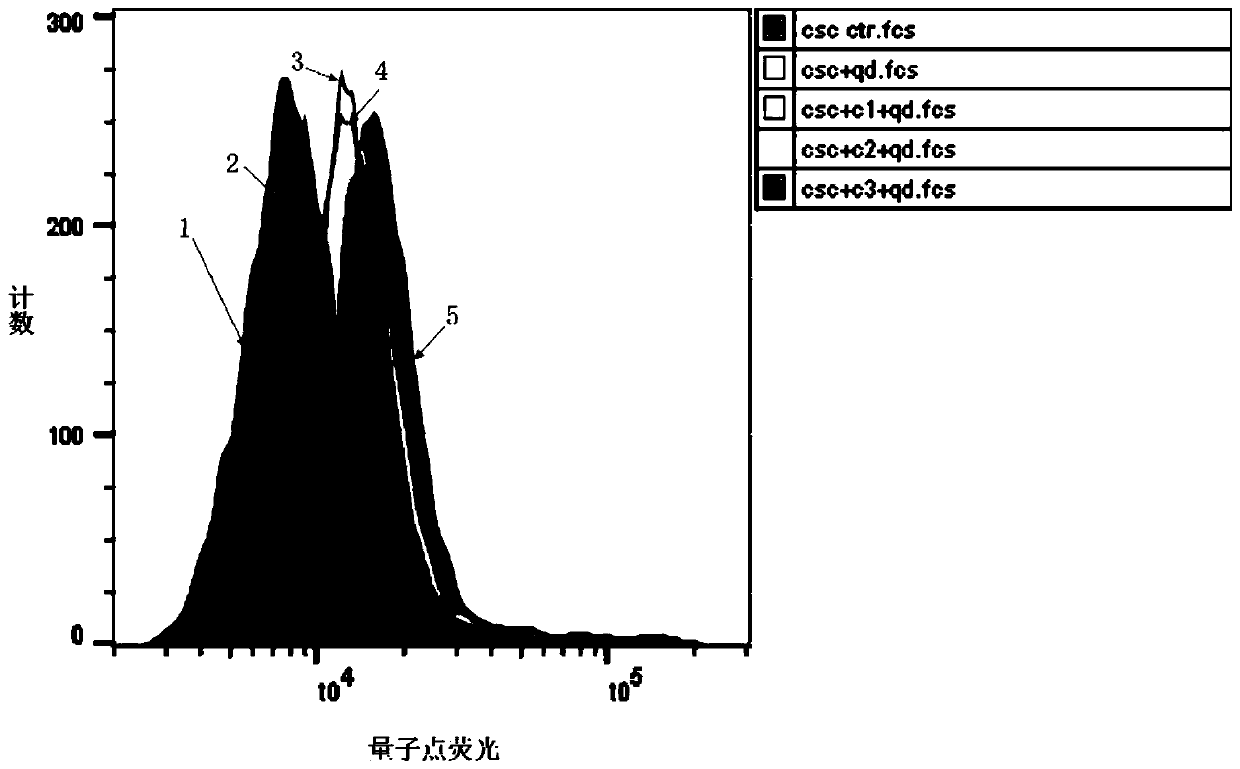
[0071] In this example, flow cytometry and surface plasmon resonance (SPR) were used to analyze the three polypeptides screened in Example 1 (the polypeptides of SEQ ID No.1 to SEQ ID No.3 are represented by polypeptides 1 to 3, respectively) Affinity and binding constants with triple-negative breast cancer stem cells, the specific steps are as follows:
[0072] a. After the three polypeptides were labeled with quantum dots, they were incubated with triple-negative breast cancer stem cells, and the fluorescence intensity was analyzed by flow cytometry to determine the affinity of the three polypeptides with triple-negative breast cancer stem cells.
[0073] b. Extract the membrane protein of triple-negative breast cancer stem cells.
[0074] c. Using EDC / NHS to couple triple-negative breast cancer stem cell membrane proteins to the SPR chip.
[0075] d. The targeting polypeptides were formulated into different concentrations, and flowed through the surface of the SPR chip in ...
Embodiment 3
[0079] Fluorescent probe targeting triple-negative breast cancer stem cells
[0080] A fluorescent probe specifically targeting triple-negative breast cancer stem cells, comprising the polypeptide whose amino acid sequence is shown in SEQ ID No.1 in Example 1 and the fluorescent group FITC coupled to the peptide chain.
[0081] To detect the targeting specificity of the fluorescent probe, the specific steps are as follows:
[0082] 1) Synthesizing a polypeptide linked to a FITC fluorescent group;
[0083] 2) Inoculate triple-negative breast cancer stem cells and MDA-MB-231 cells as a control group in a 24-well plate, and wait for the cells to grow to an appropriate density;
[0084] 3) Aspirate the medium, wash with PBS three times, each time for 3 minutes, add 500 μL of 4% paraformaldehyde solution to each well, fix at room temperature for 20 minutes, absorb the fixative, wash with PBS solution three times, each time for 3 minutes;
[0085] 4) Dilute the polypeptide at 1:50...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com