A method for synthesizing xylitol by Aspergillus oryzae engineering bacteria with enhanced saccharification ability of hemicellulose
A technology of hemicellulose and Aspergillus oryzae, applied in the field of xylitol synthesis by Aspergillus oryzae engineering bacteria, can solve the problem of low saccharification ability of hemicellulose
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
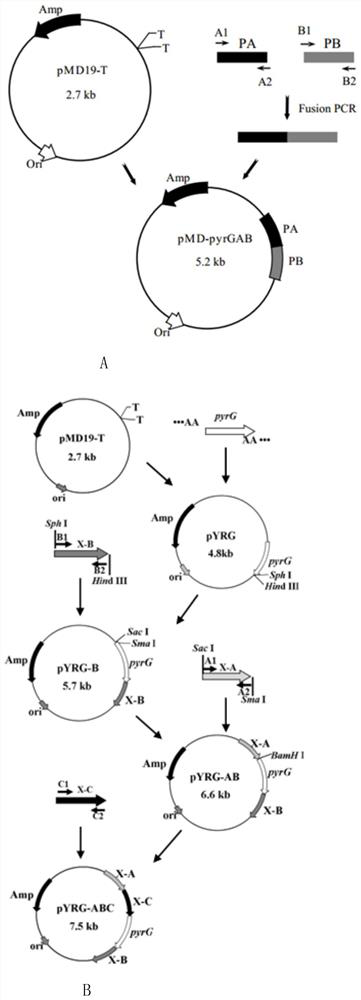
[0042] 1. Construction of pyrG knockout targeting vector pMD19-pyrGAB
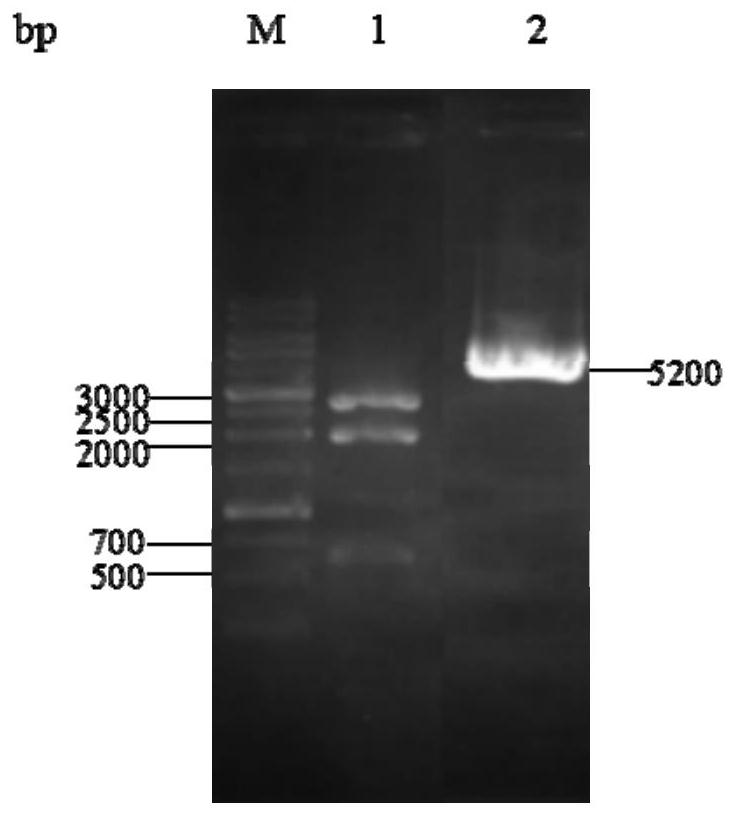
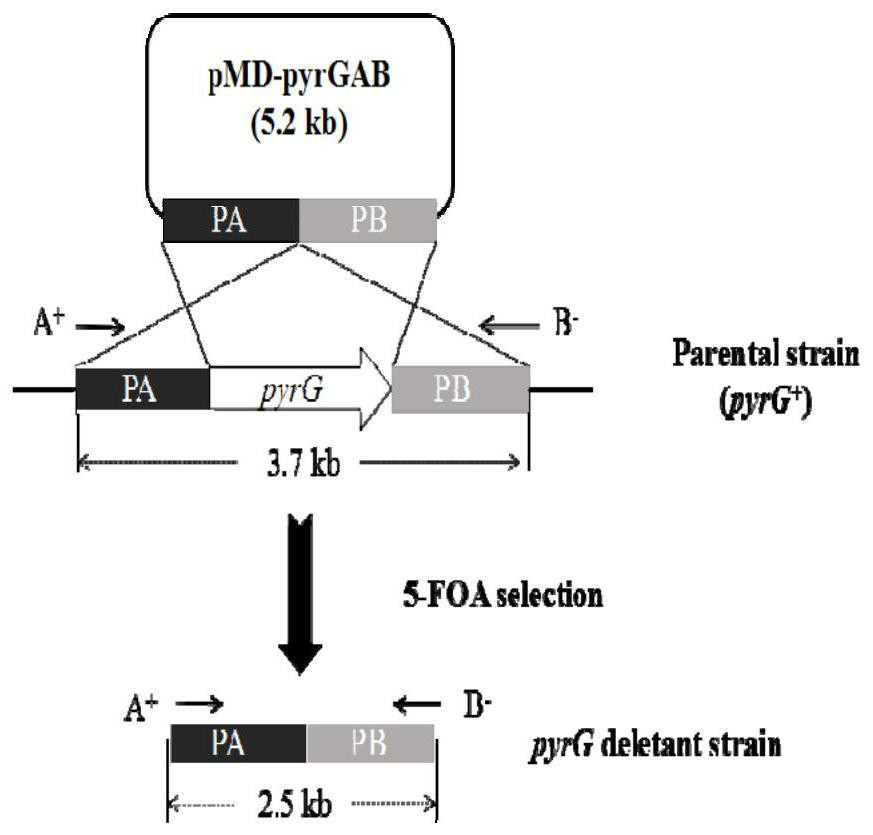
[0043] The construction process of the pyrG knockout targeting vector pMD19-pyrGAB is as follows figure 1 (A). Using the genome of A.oryzae CICC2012 as a template, the upstream and downstream homologous recombination fragments 1.0kb PA and 1.5 of pyrG (GenBank accession number: GQ496621) were amplified by PCR with primers PA-F / PA-R and PB-F / PB-R, respectively. kb PB, the upstream and downstream fragments were spliced by overlap extension PCR with primers PA-F / PB-R, and the 2.5kb spliced fragment was ligated with the 2.7kb linear vector pMD19-T to obtain a 5.2kb pyrG knockout targeting vector pMD19-pyrGAB, and Enzyme cleavage verification.
[0044] 2. In the pyrG knockout box pMD19-pyrGAB, both ends of the fusion fragment PA and PB contain Sac I and BamH I restriction sites, and the fusion fragment also contains a BamH I restriction site at about 0.6kb, so pyrG After the knockout cassette pMD-pyrGAB ...
Embodiment 2
[0052] 1. Construction of xdh seamless knockout targeting vector pMD19-pyrG-xdhABC
[0053] The construction process of the xdh seamless knockout targeting vector pMD19-pyrG-xdhABC is as follows figure 1 (B). For the positions of the upstream fragments xdh-A, xdh-B and the downstream fragment xdh-C of the Aspergillus oryzae xdh (GenBank accession number: GQ222265) gene, see Image 6 . PCR amplification of xdh-A, xdh-B, xdh-C and Four fragments of pyrG. The pyrG fragment was ligated with pMD19-T to obtain the recombinant plasmid pYRG. The recombinant plasmid pYRG-B was obtained by double digestion with restriction enzymes SphI / HindIII and ligated with the xdh-B fragment. It was digested with SacI / SmaI and ligated with the xdh-A fragment to obtain the recombinant plasmid pYRG-AB. The plasmid was digested with BamH I, and then smoothed and dephosphorylated with T4 DNAPolymerase and Alkaline Phosphatase, respectively, and blunt-end ligated with the xdh-C fragment treated wit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com