Eml4-alk gene mutation analysis method
An analysis method and gene technology, applied in the field of EML4-ALK, can solve the problems of cost and time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1: Preparation of high-density microchips
[0048] For the high-density microchip used in the experiment, the pore-size is formed from 5.5 to 8.5 μm, and white blood cell (WBC) and red blood cell (RBC) are smaller than 5.5 μm. The chip is removed by passing it through, and the target cells with a size greater than 5.5 μm are microchips developed in a way that they can be prepared into a chip to selectively recover a specific size.
[0049] For reference, in order to confirm the cell recovery rate of high-density microchips, 10, 100, and 1000 cancer cells of cancer patients were spiked through the chip to perform the recovery of cancer cells on the observation chip. Rate of experiment. The results are shown in Table 1 below.
[0050] Table 1
[0051] Sample Number of spiked cells Number of recovered cells Cell recovery rate (%) 110990 21008686 3100085085
[0052] Calculating the cell recovery rate of the chip, the result showed a high cell recovery rate of about 8...
Embodiment 2
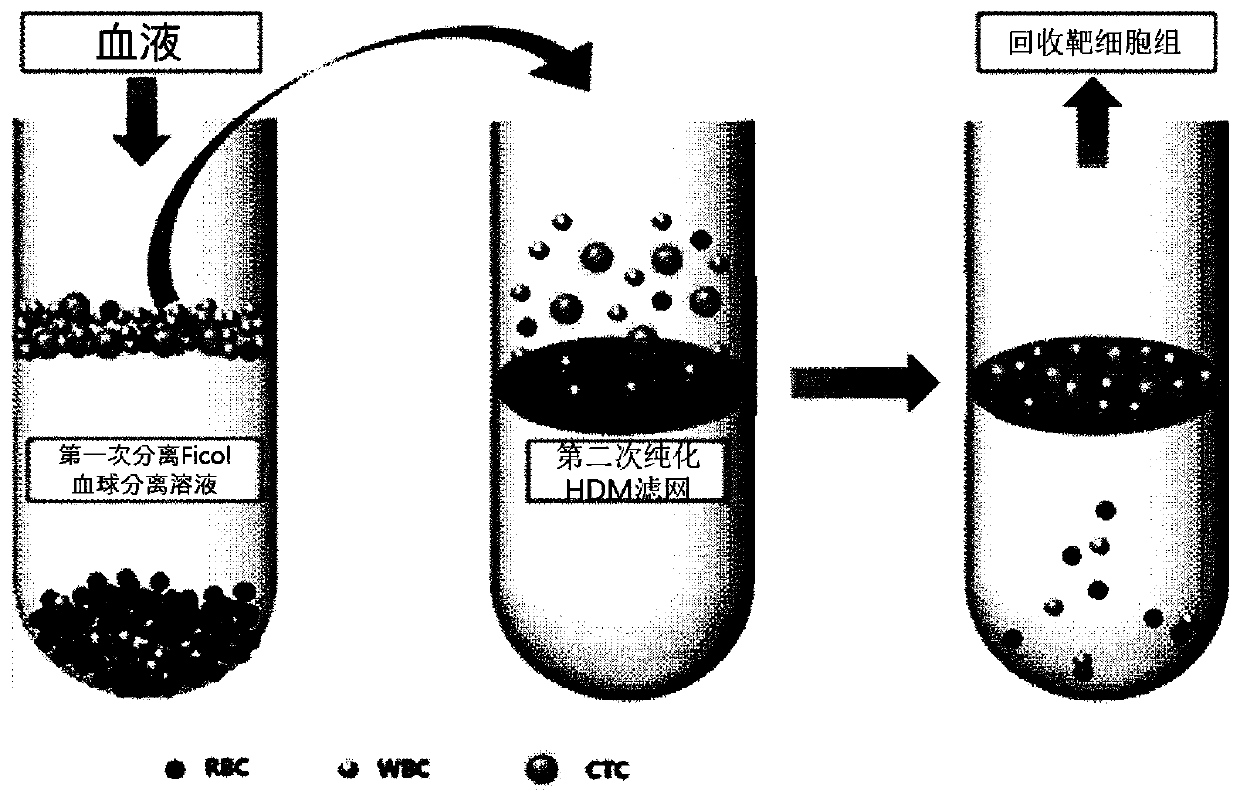
[0053] Example 2: Separation process of circulating cancer cells (CTC) in blood
[0054] 1. Put 250μl of antibody polymer in 5ml of blood, mix for about 3 seconds, and react for 20 minutes at room temperature.
[0055] 2. Add 5 ml of phosphate buffered saline (PBS) containing 1% fetal bovine serum (FBS).
[0056] 3. Carefully put 10 ml of the reaction solution in a 50 ml tube containing 15 ml of Ficoll solution.
[0057] 4. Centrifuge the solution at 1200g for 20 minutes to remove blood cells for the first time.
[0058] 5. In order to prevent the adsorption of unnecessary cells, a high-density microchip (HD Microporous chip, filter) is treated with 0.1% bovine serum albumin solution for 10 minutes and coated, and then washed with phosphate buffer.
[0059] 6. Put the supernatant solution of Ficoll on the filter, and gravity filter the red blood cells present in a small amount to separate high-purity circulating cancer cells in the blood for the second time. It does not perform centrif...
Embodiment 3
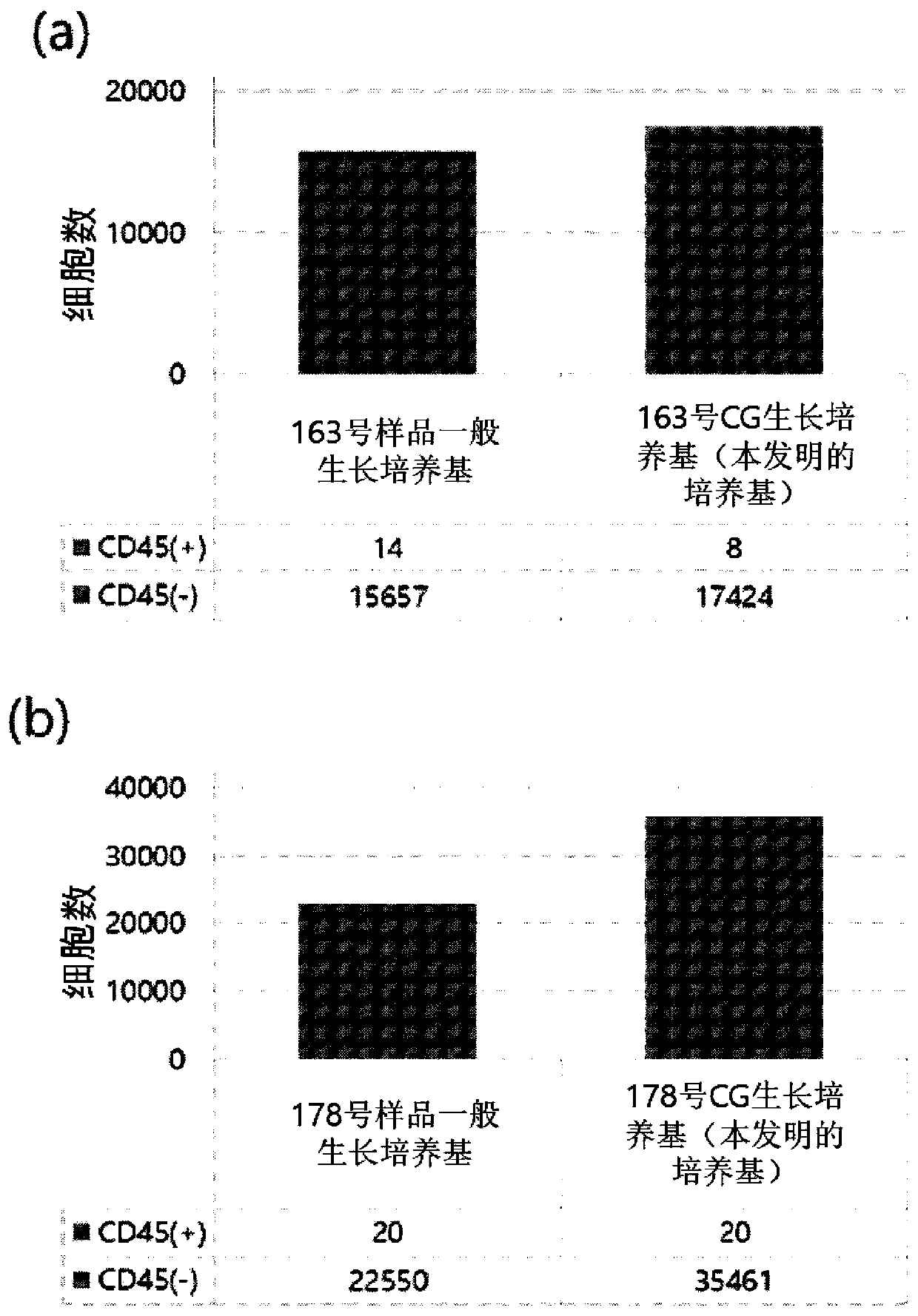
[0061] Example 3: Short-term culture of circulating cancer cells in isolated blood
[0062] Circulating cancer cells in the blood separated by the high-density microchip of the present invention are seeded in an ultra low attachment culture coated with a hydrogel with neutral charge and hydrophilicity. board. The above-mentioned culture plate contains a culture medium containing 11 ng / ml insulin, 22 ng / ml transferrin, 2 ng / ml EGF and 8 μm Roche kinase inhibitor. For the above-mentioned short-term culture, from the time point of culture to 14 During the day, in a cell incubator at 37℃ and 5-10% CO 2 Culture under the conditions.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com