DNA library construction kit based on illumina sequencing platform, library construction method and application
A DNA library and library construction technology, applied in the field of DNA library construction kits, can solve the problems of low amplification efficiency, high price of finished kits, low success rate of library construction, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
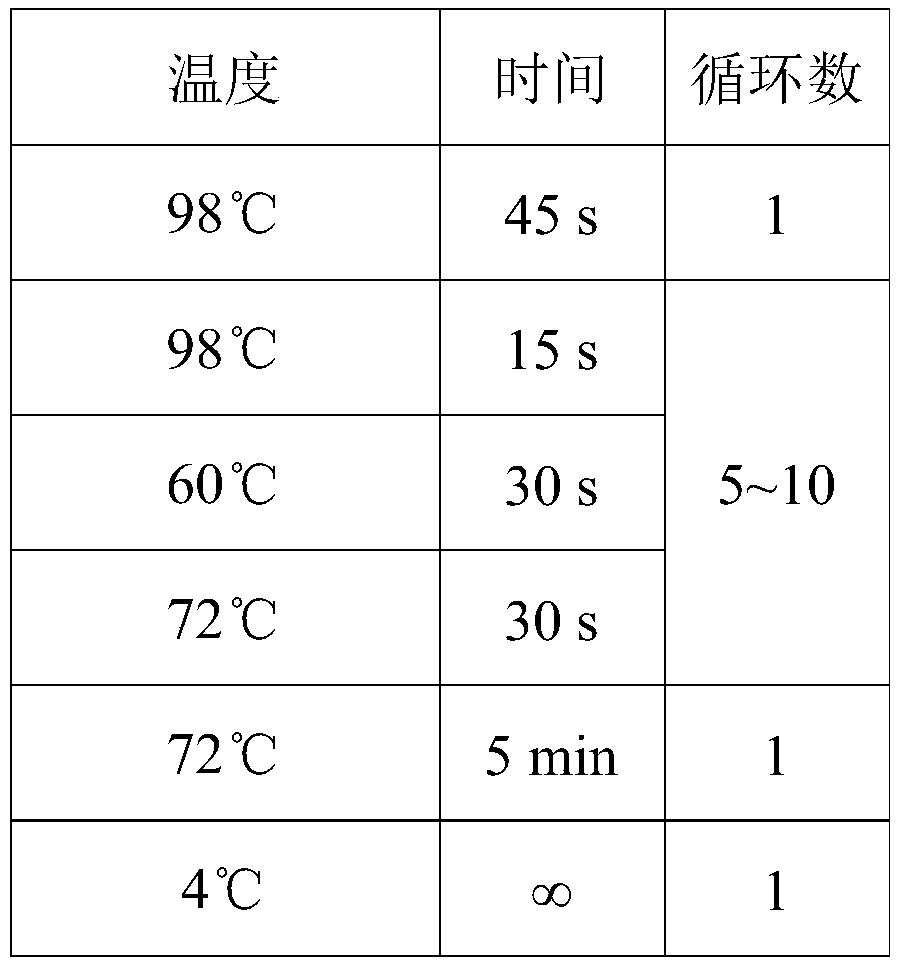
[0077] Example 1 Genomic DNA Fragmentation
[0078] In this example, after using Qubit3.0 to quantify genomic DNA, add 300ng of sample into a 0.2mL PCR tube, add 0.1×TE buffer to make up to 55μL, mark the sample number on the tube cap, mix well and centrifuge; open Ultrasonic cleaner, set the ultrasonic time of the ultrasonic cleaner to 5min, and place 0.2mL PCR tubes containing samples in order around the ultrasonic sounding position, and interrupt 9 samples at a time; after ensuring that the PCR tube is clamped, inject pure water to the ultrasonic wave. At the 2 / 3 scale of the cleaning instrument, click the start button to perform ultrasound interruption. The parameter settings are shown in Table 1. After the ultrasound is completed, the fragmented DNA is purified with AMpure XP magnetic beads and dissolved in 39.5 μL NF water.
[0079] Table 1 Ultrasonic treatment parameters
[0080] Expected fragment length (bp) Power (W) temperature(℃) Ultrasonic time (s) ...
Embodiment 2
[0081] Example 2 End Repair and Add A Reaction
[0082] Prepare the end-repair reaction solution according to Table 2 to complete the end-repair and A addition reaction of fragmented DNA; blow and mix with a pipette, collect the reaction solution to the bottom of the tube after brief centrifugation, and perform end-repair and A addition according to the conditions in Table 3. A response.
[0083] Table 2 End repair and A reaction system
[0084] Reagent Volume (μL) Fragmented DNA 37 end repair buffer 10 end repair enzyme 3
[0085] Table 3 End Repair and Add A Reaction Conditions
[0086] temperature Reaction time 22℃ 10min 72℃ 10min 4℃ ∞
Embodiment 3
[0087] Embodiment 3 linker ligation reaction
[0088] Prepare the adapter ligation reaction solution according to Table 4 to complete the adapter ligation of the fragmented DNA, wherein the adapter set working solution is self-synthesized P5 (SEQ ID NO: 6-10) and P7 adapter sequence (SEQ ID NO: 6-10) with a molar ratio of 1:1. ID NO: 1-5), the mixed sequence diluted to 10 μM; use a pipette to mix well, centrifuge briefly to collect the reaction solution to the bottom of the tube, and perform the adapter ligation reaction according to Table 5.
[0089] Table 4 Adapter Ligation Reaction System
[0090] Reagent Volume (μL) end repair plus A product 50 ligation buffer 10 T4 DNA ligase 2 Linker set working solution (10μM) 3
[0091] Table 5 Adapter Ligation Reaction Conditions
[0092] temperature Reaction time 22℃ 15min 4℃ ∞
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com