Prader-Willi Syndrome detection kit and use method thereof
A detection kit and technology for Wiley syndrome, applied in the field of diagnosis and research of chubby Wiley syndrome, can solve the problems of difficult detection of microdeletions, large sample input, complicated operation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0093] Example 1 Rapid extraction of nucleic acid from clinical samples (such as blood, saliva, urine)
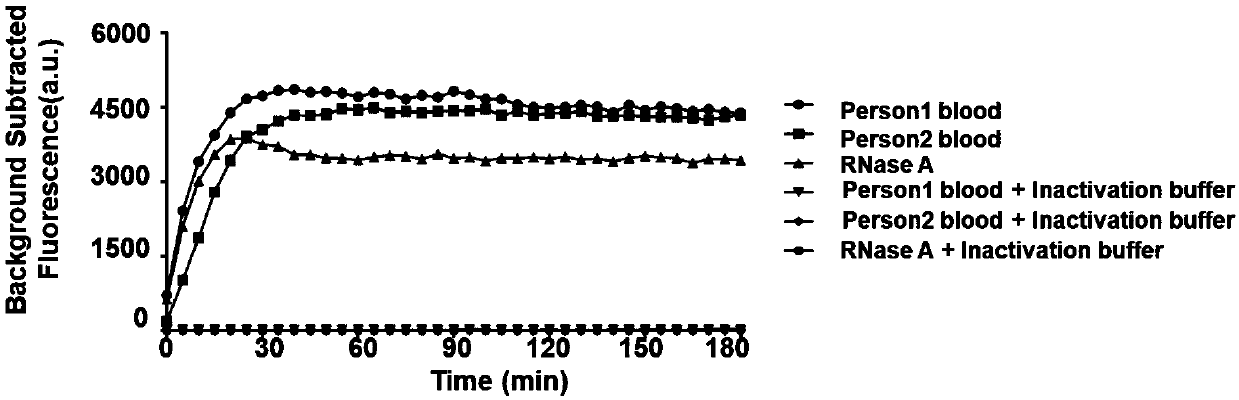
[0094] In order to realize rapid extraction of nucleic acid, the inventors added an RNase inactivator to the sample, and then lysed the cells at 95° to release the nucleic acid. The specific steps are as follows, add RNase inactivator (TCEP / EDTA with a final concentration of 100mM / 1mM) to the sample (such as blood, saliva, urine), pipette and mix with a sterile pipette tip (Thermo QSP), 95° Heat for 10 minutes, centrifuge at 14000 rpm for 10 minutes, and take the supernatant. RNase activity in the supernatant was detected with RNaseAlert Lab Test Kit v2 (Thermo), and the real-time fluorescence change was monitored with a multifunctional microplate detector (BioTek SynergyNEO) (such as figure 1 shown), the RNase inactivator can effectively inhibit the RNase activity in the sample (blood), thereby inhibiting its degradation of RNA, effectively protecting the RNA to be detect...
Embodiment 2
[0095] Example 2 Screening for efficient and specific primers for sno-lncRNAs molecular markers for recombinase polymerase amplification
[0096] 2.1 Design and synthesis of specific recombinase polymerase amplification primers
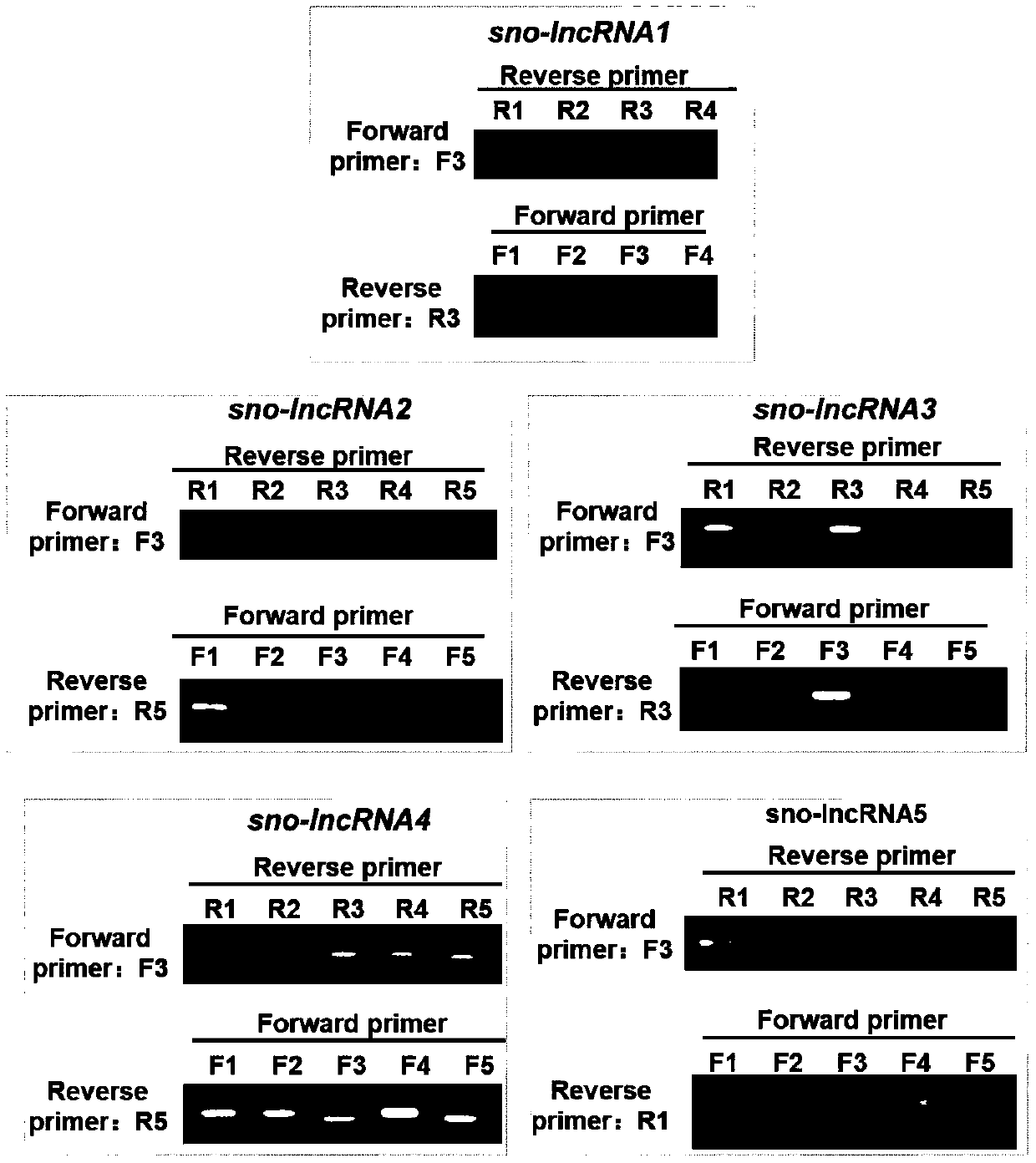
[0097] The sno-lncRNAs molecular marker combination of chubby-Willi syndrome includes 5 long noncoding RNAs, namely sno-lncRNA1, sno-lncRNA2, sno-lncRNA3, sno-lncRNA4, and sno-lncRNA5. Sequences are in the NCBI GeneExpression Omnibus database ( https: / / www.ncbi.nlm.nih.gov / geo / ; accession number GSE38541). In order to realize trace detection, the present invention utilizes recombinase polymerase to amplify sno-lncRNAs molecular markers. The recombinase polymerase amplification primers of the chubby-Willi syndrome sno-lncRNAs molecular marker combination are multiple pairs of specific primers, and the primers are designed through the NICB Primer-BLAST online primer design tool ( https: / / www.ncbi.nlm.nih.gov / tools / primer-blast / ), and the prime...
Embodiment 3
[0113] Example 3 Screening gRNA (CRISPR guide RNA) for efficient and specific targeting of sno-lncRNAs molecular markers
[0114] The nucleotide sequence of the gRNA is as follows:
[0115] Table 2
[0116]
[0117]
[0118]
[0119]
[0120]
[0121]
[0122]
[0123]
[0124]
[0125]
[0126]
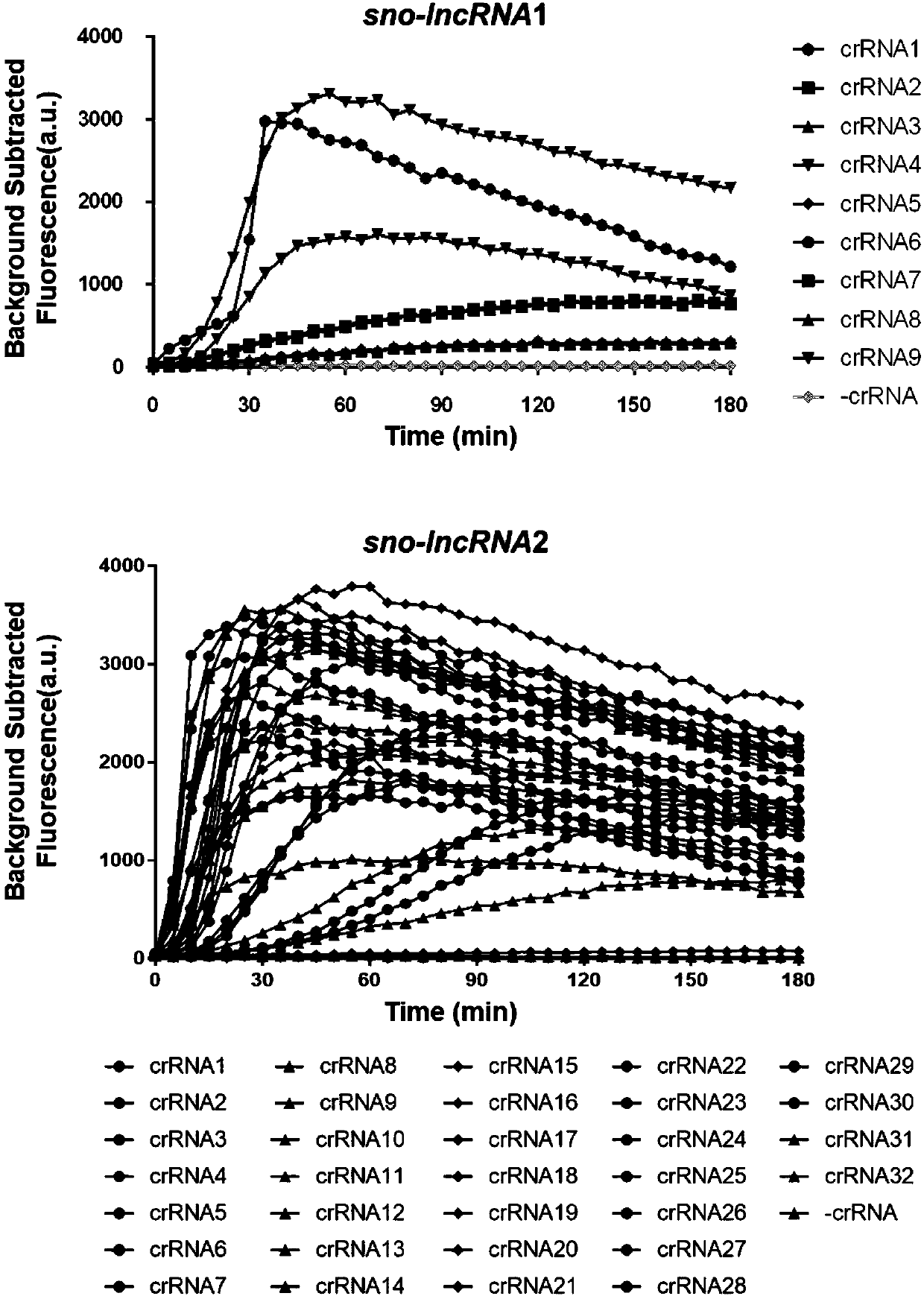
[0127] Screen gRNAs that are highly efficient and specifically target molecular markers of sno-lncRNAs:
[0128] Under the "guidance" of gRNA, Cas13a binds to the target RNA to exert its cutting activity, and the "activated" Cas13a will not only cut the target RNA, but also non-specifically cut other RNAs. Therefore, gRNAs with high efficiency were screened by the ability of Cas13 to cleave fluorescent RNA substrates. The fluorescent RNA substrate RNAse Alert v2 was purchased from a company (Integrated DNA Technologies). Cas13 cutting RNA reaction system is as follows:
[0129]
[0130]
[0131] 37 ° reaction, 3h, real-time monitoring substr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com