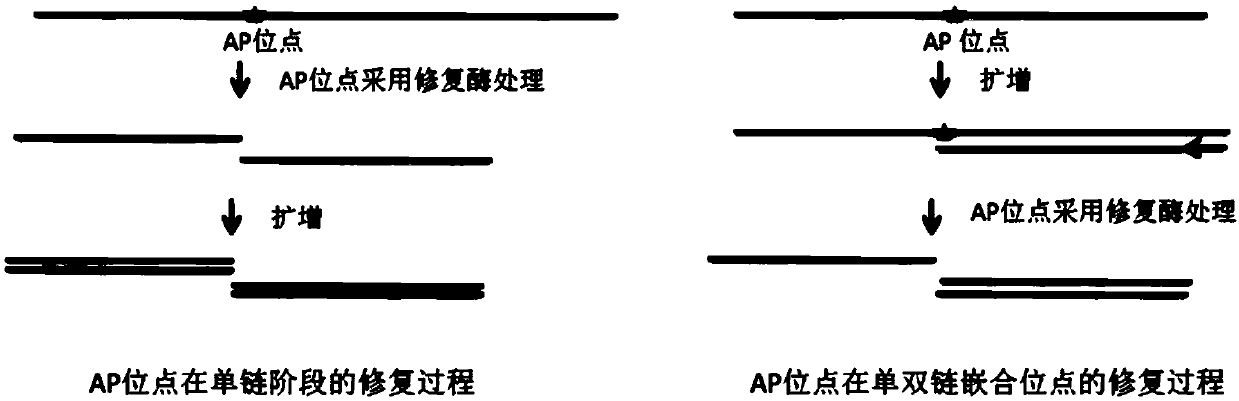
Single-chain library building method of whole-genome methylated library and obtained whole-genome methylated library
A genome-wide, methylation technology, applied in the field of genome methylation research, can solve the problems of waste, too much sequencing data, low efficiency, etc., and achieve the effect of reducing the cost of library construction and improving the coverage.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
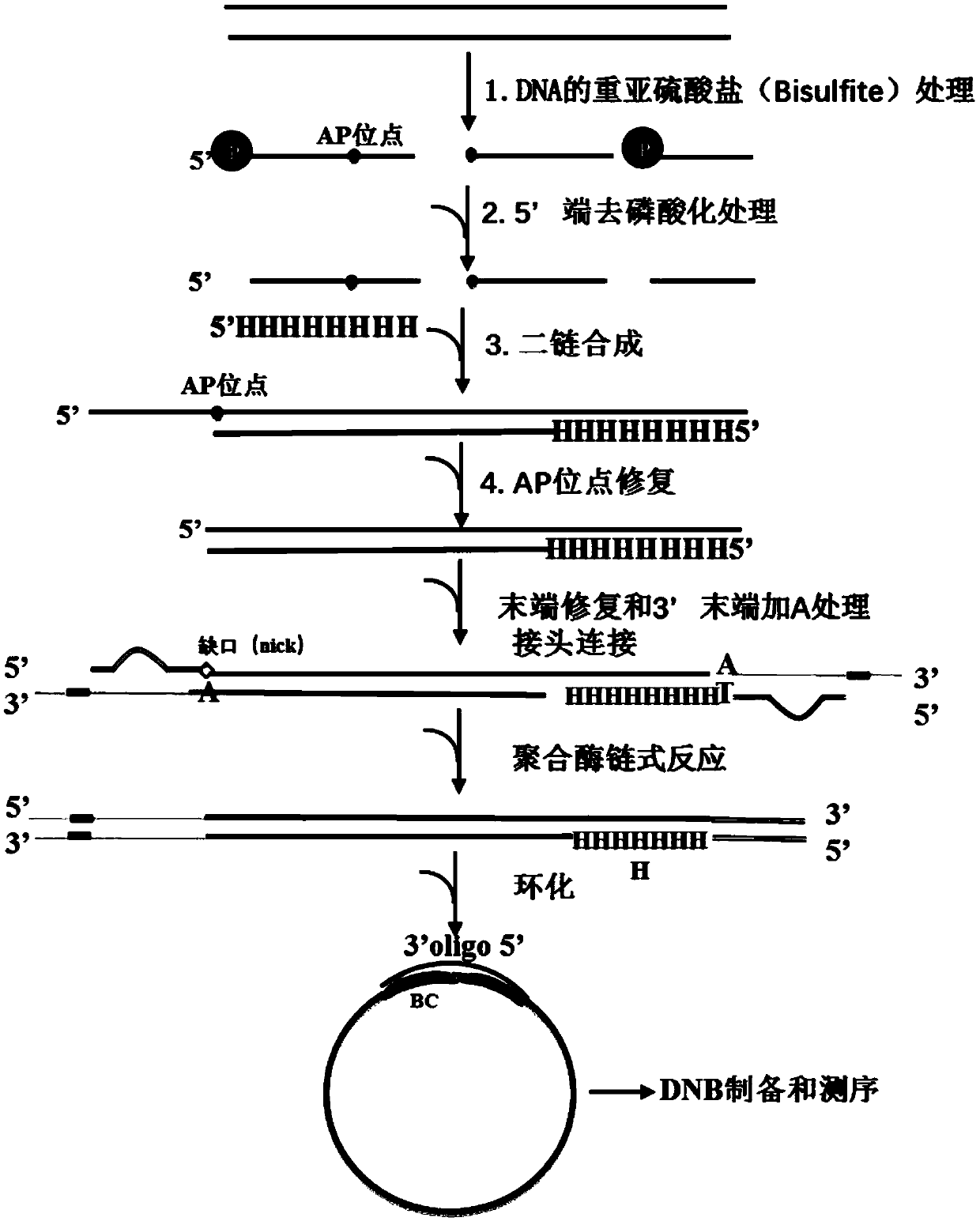
[0054] The sample in this embodiment is unfragmented genomic DNA.
[0055] (1) Bisulfite treatment of DNA:
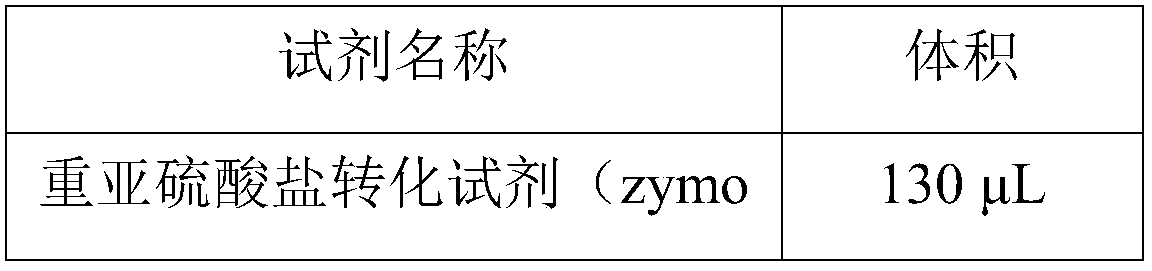
[0056] Prepare the reaction mixture shown in Table 1 below:
[0057] Table 1
[0058]
[0059]
[0060] The reaction conditions are shown in Table 2 below:
[0061] Table 2
[0062] Bisulfite Treatment Conditions time 98℃ 10min 64℃ 2.5h 4℃ ∞
[0063] After the reaction, purify with a purification column and dissolve back in 10 μL.
[0064] (2) 5' end dephosphorylation treatment:
[0065] Prepare the reaction mixture shown in Table 3 below:
[0066] table 3
[0067]
[0068] The reaction conditions are shown in Table 4 below:
[0069] Table 4
[0070]
[0071]
[0072] After the reaction, place on ice for 3-5 minutes.
[0073] (3) Two-strand synthesis reaction:
[0074] Prepare the reaction mixture shown in Table 5 below:
[0075] table 5
[0076] Reagent volume 10X NEB2 buffer 3μL 2.5mM dN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com