Primer group, kit and method for simultaneously detecting pigeon circovirus, pigeon adenovirus and pigeon herpes virus type 1
A pigeon circovirus and pigeon herpes virus technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of low isolation rate, easy to cause missed diagnosis, long cycle, etc., to achieve High sensitivity, reduced labor costs, high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1: Extraction of viral DNA
[0042] Scrape the mucous membrane of the oral cavity, sinuses, and esophagus to be tested, and put 10-20mg of the sample into a 1.5mL centrifuge tube. Centrifuge at 12,000×g for 3 minutes, and transfer the supernatant to a new 1.5mL centrifuge tube. Add 230 μL BufferGA and 20 μL PK working solution, and vortex to mix. Water bath at 55°C until the tissue is completely enzymatically hydrolyzed. Add 250μl BufferGB to the digestion solution, vortex mix at the highest speed for 20sec, and bathe in 70℃ water for 10min. Then column purification: add 250 μL absolute ethanol to the digestion solution, vortex and mix for 15-20sec. Put the gDNA Columns adsorption column into the Collection Tubes 2mL collection tube. Transfer the mixed solution (including the precipitate) obtained in the previous step to the adsorption column. Centrifuge at 12,000×g for 1 min. Discard the filtrate and place the adsorption column in a collection tube. Add ...
Embodiment 2
[0044] Example 2: Design and synthesis of primers and effectiveness testing
[0045] 1. Design and synthesis of primers: based on Hexon gene of PiAdV (GenBank: MF576429.1), Rep and capsid gene of PiHV-1 (GenBank: JX901125.1) and DNA polymerase of PiCV (GenBank: KJ995972.1) in Genbank Gene nucleic acid sequences were compared with DNA-Star software, and conserved regions were screened out. Specific primers were designed with Primer5.0 software, and then verified with NCBI BLAST. The following three pairs of primers were screened out, as shown in Table 1.
[0046] Table 1 is used to detect the primer design sequence of three kinds of pathogens
[0047]
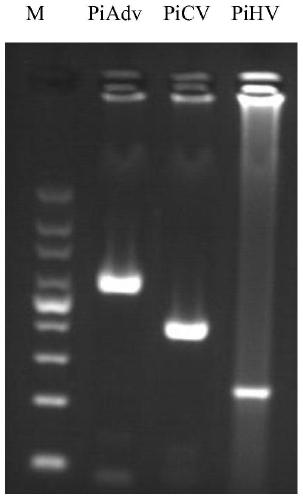
[0048] Among them, the predicted amplified fragment sizes are: PiAdV product is 616bp, PiHV-1 product is 239bp and PiCV product is 418bp. Primers were synthesized by Nanjing Sipujin Biotechnology Co., Ltd.
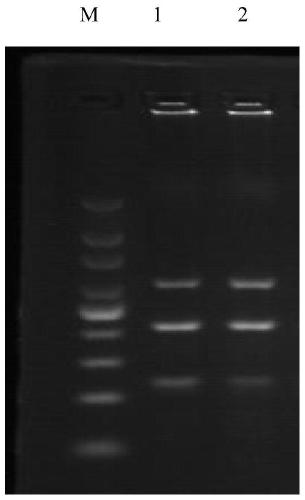
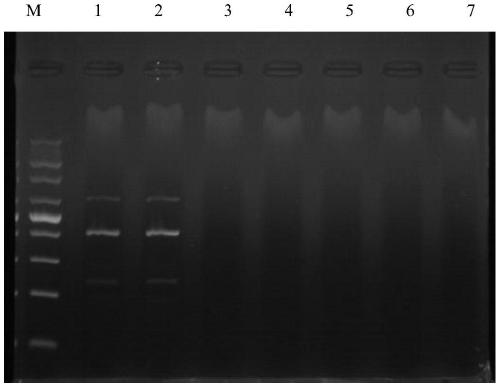
[0049] 2. Effectiveness detection: single PCR detection and triple PCR detection of clinical samples
[0050] (1) A ...
Embodiment 3
[0066] Embodiment 3: kit and detection method
[0067] 1. The composition of this kit: primers for amplifying PiAdV, PiHV-1 and PiCV; 2×Es Taq MasterMix; double distilled water; positive DNA templates for PiAdV, PiHV-1 and PiCV; DNA genome extraction kit (purchased from Nanjing Novizan Biotechnology Co., Ltd., model DC102); PCR reaction reagents and electrophoresis detection reagents.
[0068] 2. The content of each component:
[0069] (1) Primers: Primers were synthesized by Nanjing Sipujin Biotechnology Co., Ltd. The total amount of each primer for amplifying PiAdV, PiHV-1, and PiCV is 2OD when synthesized. Divide 1OD into 1 tube, then add double-distilled water to dissolve it to 10mM, and then pack into 100μL tube, freeze at -20℃, Grab and go.
[0070] (2) DNA genome extraction kit (purchased from Nanjing Novizan Biotechnology Co., Ltd., model (DC102-01),
[0071] ① Product use: This product is suitable for animal tissues from 1-20mg and less than 5×10 6 Extract genomi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com