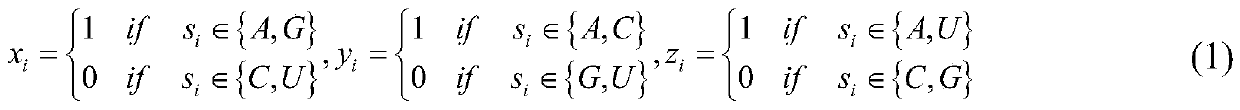Method for predicting N6-methyladenosine modification site in RNA based on stacking integration
A technology of methyladenosine and prediction methods, applied in the field of systems biology, can solve problems such as single prediction model, single species, and limitations of traditional classifiers, and achieve good prediction performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0033] In order to make the object, technical solution and advantages of the present invention clearer, the invention will be further described in detail below in conjunction with the accompanying drawings and embodiments. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
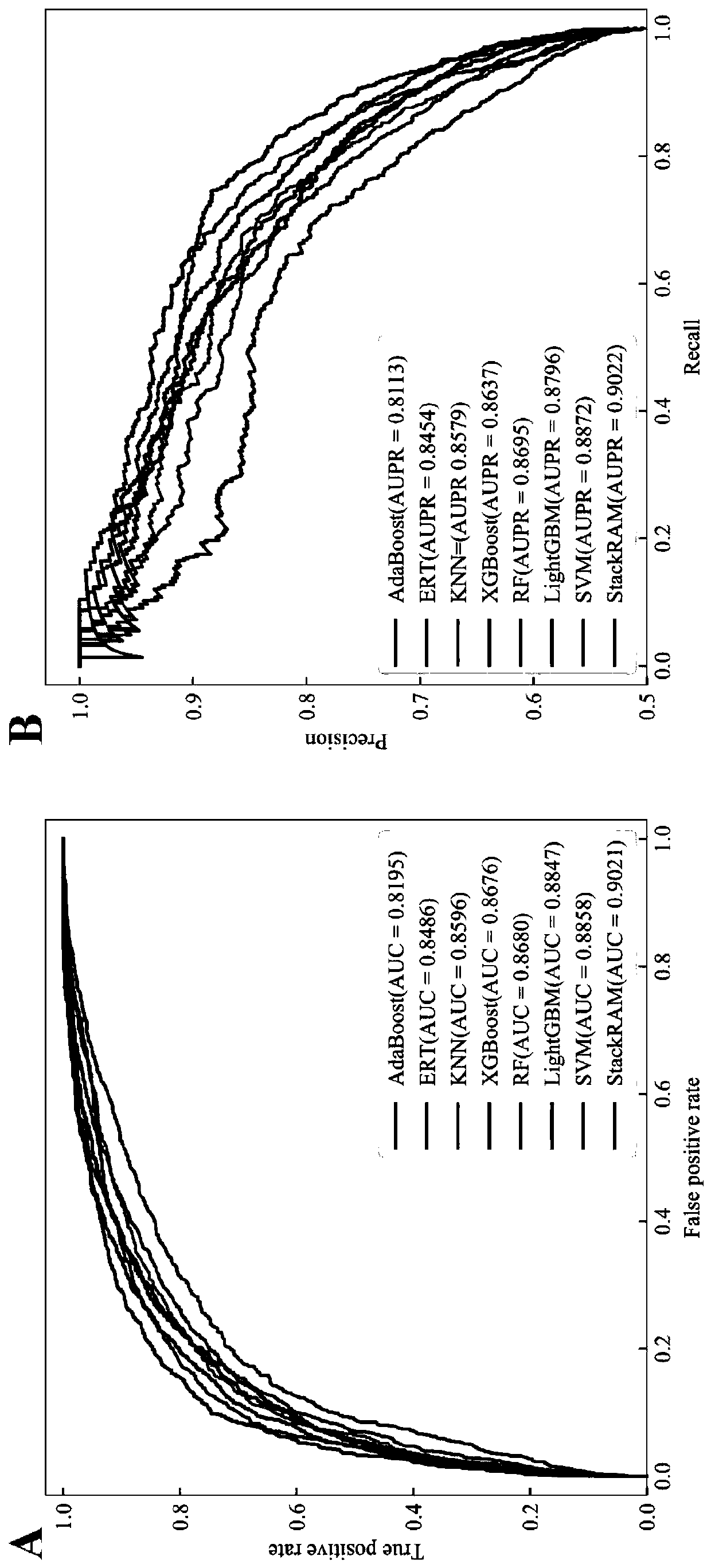
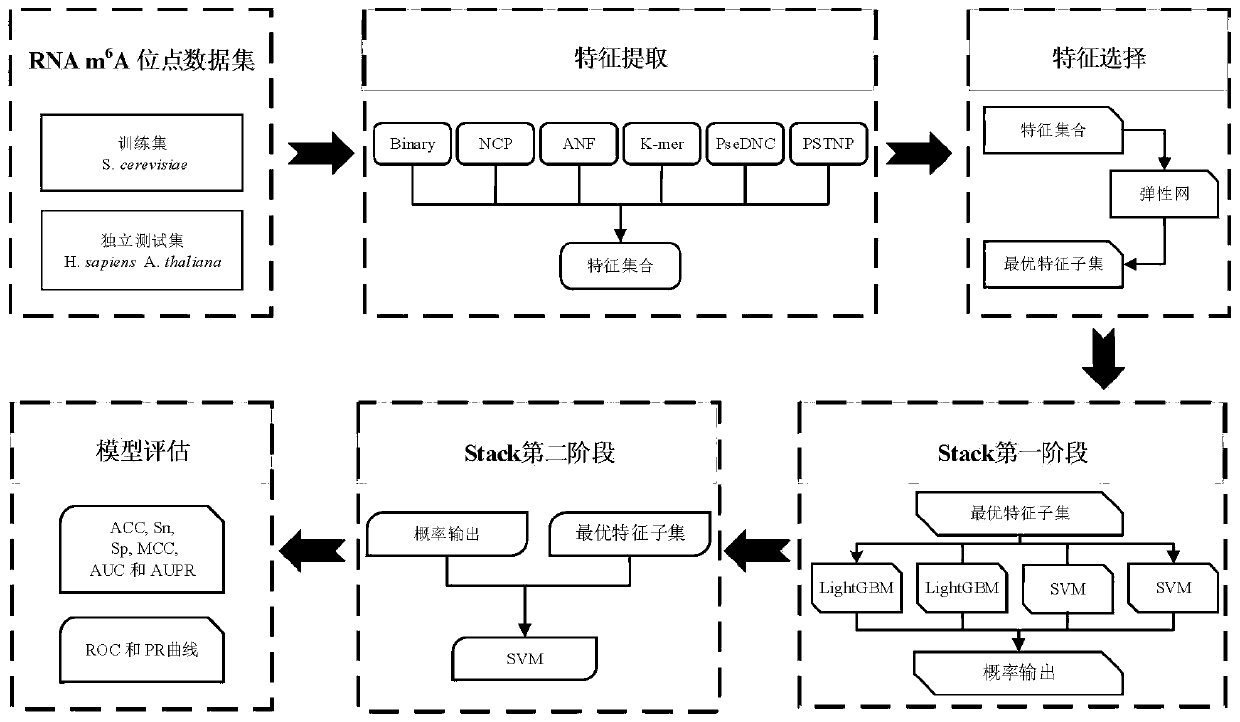
[0034] A stacking-based integration of m in different species of RNA 6 A site prediction methods, such as figure 1 shown, including the following steps:
[0035] 1) Collect N in RNA 6 -Methyladenosine modification (m 6 A) Site information: Get the N of RNA from 3 different species 6 -Methyladenosine modification site data set, including the RNA sample sequence of the positive and negative data set and the corresponding category label.
[0036] 1-1) The real data of three species of S.cerevisiae, H.sapiens and A.thaliana are selected; the data set S.cerevisiae is used as a training set, and the data se...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com