Single nucleotide polymorphism (SNP) marker related to growth speed of basa fish and application of SNP marker
A growth rate, pangasius technology, which is applied in the determination/inspection of microorganisms, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problem of molecular markers to be excavated, etc., and achieve low-cost results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1 Obtaining of SNP markers related to Pangasius growth rate
[0032] 1.1 Acquisition of pangasius populations
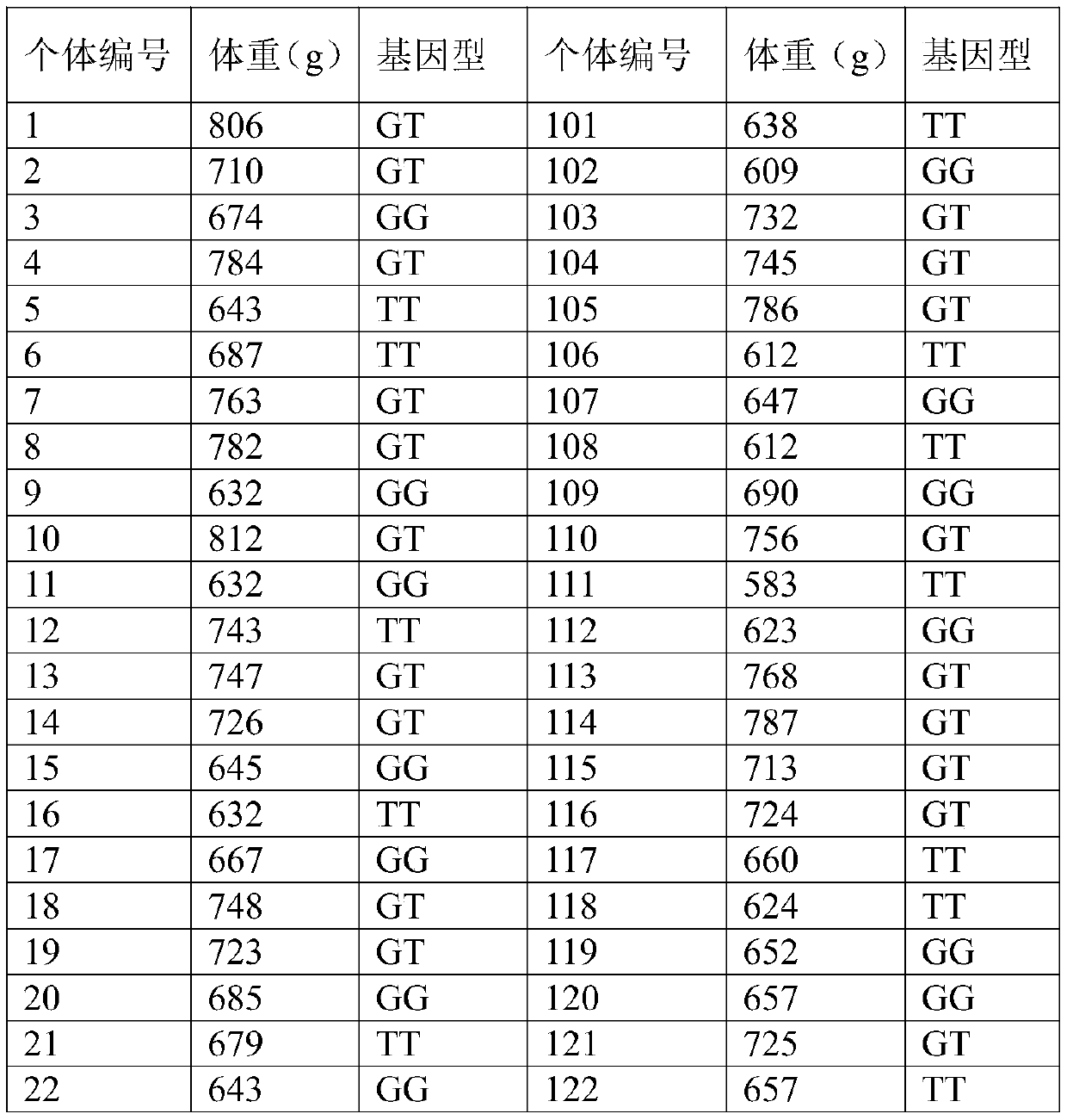
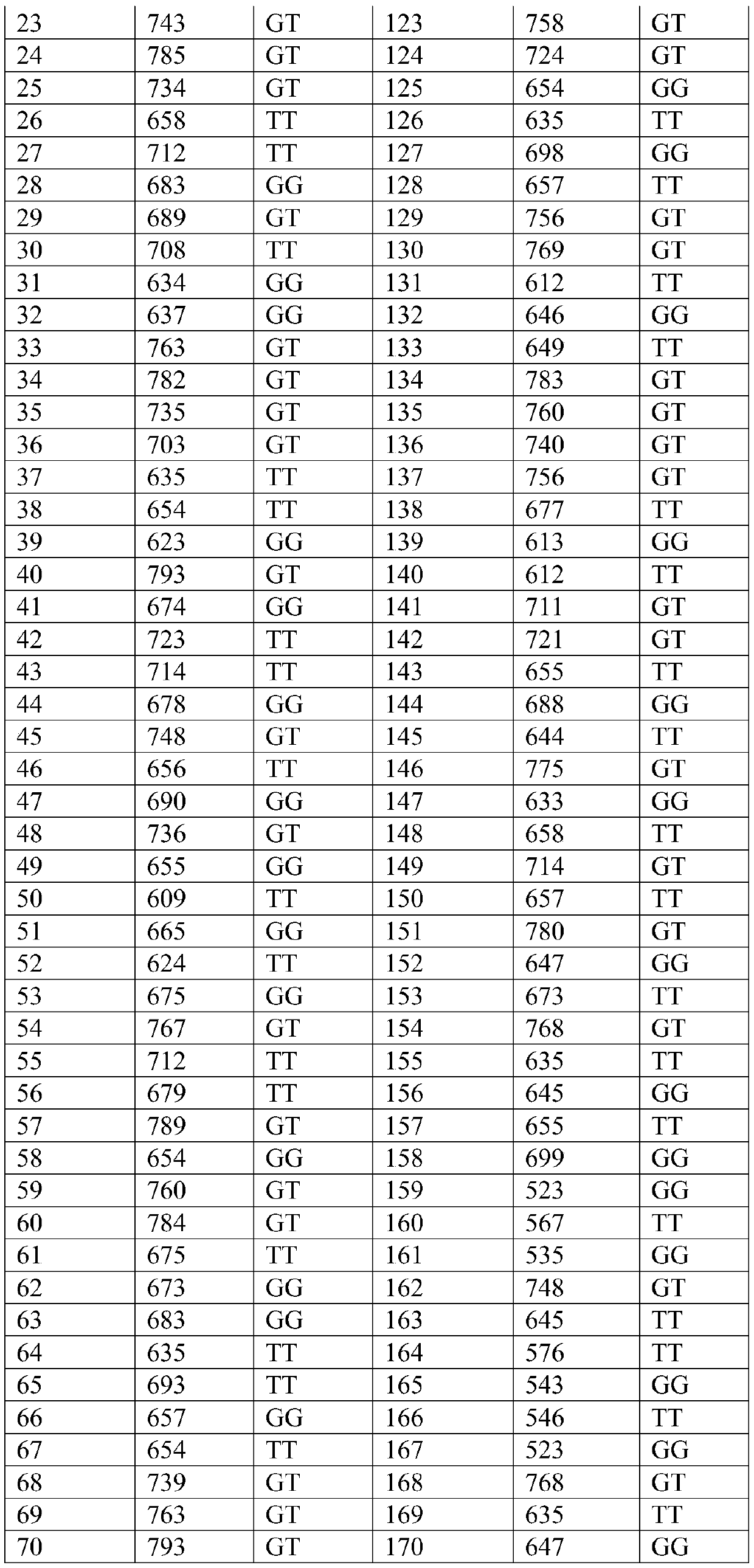
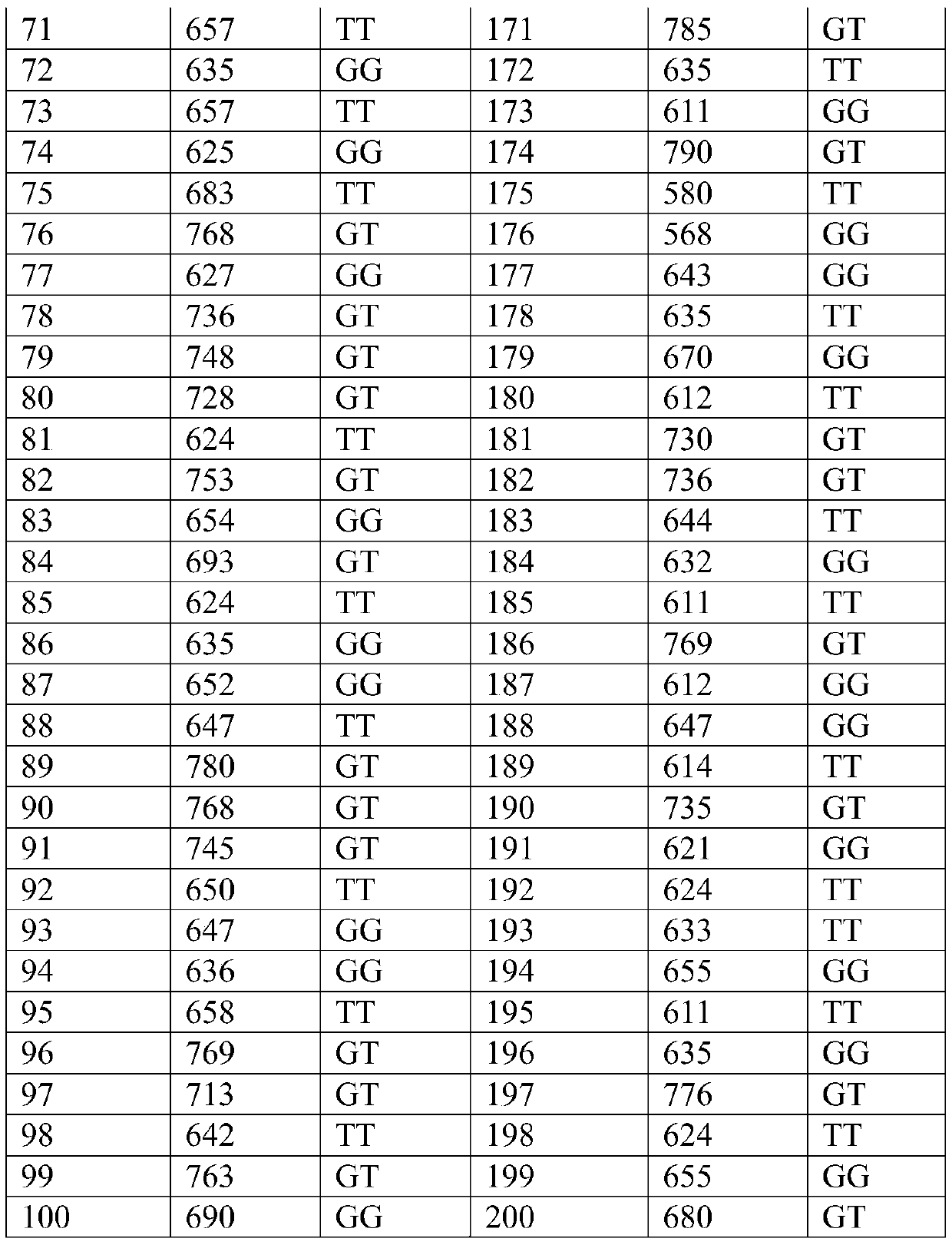
[0033] The group used was pangasius hatched on June 15, 2017 in a pangasius farm in Lingshui, Hainan. On August 2, 2017, 60,000 fry were transferred to a 10-acre pond for further breeding. On May 8, 2018, 500 individuals were randomly selected from the pond, and the dorsal fin rays of the fish were cut and stored in 95% ethanol at -20°C for genomic DNA extraction.
[0034] 1.2 Pangasius genomic DNA extraction
[0035] In this experiment, the conventional phenol-chloroform method was used to extract the genomic DNA in the fin rays of pangasius, and the specific steps were as follows:
[0036] (1) Take 0.3-0.5g of fin rays in a 1.5ml Eppendorf tube, cut it into pieces, and dry it for 20 minutes on a clean bench;
[0037] (2) After the ethanol is basically volatilized, wash with TE buffer solution (10mmol / ml Tris, 1mmol / ml EDTA, SDS 5%, pH=8.0) for ...
Embodiment 2
[0042] Example 2 Sequencing verification and application of SNP markers related to pangasius growth rate
[0043] 2.1 Extract the genomic DNA in the fin rays of pangasius to be tested
[0044] The pangasius to be tested came from the pangasius group in Example 1, and 200 fish were randomly selected again, and the genomic DNA was extracted according to the DNA extraction method described in Example 1.
[0045] 2.2 Amplification of nucleotide fragments containing SNP sites
[0046] Using the genomic DNA of each pangasius to be tested obtained by the aforementioned extraction as a template, the forward primer F: 5'-CTCCTTGTGTGCGTCCTGGTGTGTCTAA-3' (SEQ ID NO: 2) and the reverse primer R: 5'-AAAACATTGCTGTTTGTTATTTAAAA-3'( SEQ ID NO: 3), amplifying the nucleotide fragment where the SNP to be detected is located. Among them, the PCR reaction system is calculated as 25 μl: 50-100ng / μl template DNA 1μl, 10pmol / μl primer F and R 1μl each, 10mmol / L dNTP mix 2.0μl, 5U / μl Taq DNA polymer...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com