Reagent kit for detecting novel coronaviruses and influenza viruses
A technology for coronaviruses and influenza viruses, applied in the field of molecular biology detection, to achieve good specificity, avoid false positives or false negatives, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
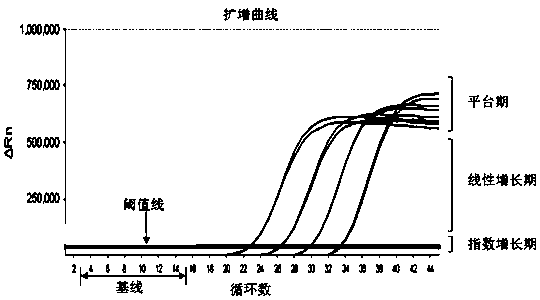
Image
Examples
Embodiment 1
[0062] Embodiment 1 Primer probe, reagent and detection method
[0063] 1.1 Primer probe sequence
[0064] The ORF1ab gene of the novel coronavirus COVID-19 was selected as the amplification target region, specific primers and fluorescent probes were designed, and the probes were labeled with FAM to detect whether the samples contained novel coronavirus RNA. Select the S gene of the new coronavirus, design specific primers and fluorescent probes, and the probes are labeled with ROX to detect whether the sample contains the new coronavirus RNA. The characteristic conserved sequence of influenza virus A was selected, specific primers and fluorescent probes were designed, and the probe was labeled with Cy5 to detect whether the sample contained influenza virus ARNA. The characteristic conserved sequence of influenza virus B was selected, specific primers and fluorescent probes were designed, and the probe was labeled with VIC to detect whether the sample contained influenza viru...
Embodiment 2
[0095] Embodiment 2 detection sensitivity
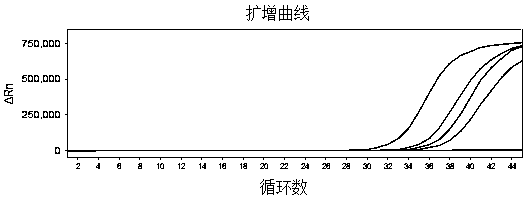
[0096] The sensitivity was verified using the primer-probe combination (Table 2), reagents, and nucleic acid amplification detection method in Example 1.
[0097] For COVID-19, the RNA extracted from 3 clinical real samples was mixed into 1 part. After quantification by ddPCR, it was 6460copeis / μL, and the samples were diluted to 8 copies / μL, 0.8copies / μL, and 0.4copies / μL with virus preservation solution. μL, 0.2copies / μL, 0.1 copies / μL, 0.05 copies / μL, for nucleic acid extraction and amplification.
[0098] S4 (S4:inFluA / H3N2, 1.2×10 4 copies / μL) were evaluated, and the virus preservation solution was used to perform 10-fold serial dilutions, and were diluted to 1.2×10 3 Copies / μL, 120copies / μL, 12copies / μL, 1.2copies / μL, 0.12copies / μL, amplified after extraction.
[0099] Influenza virus B (influB) uses S1 of the national reference product of the second-generation influenza A / B nucleic acid detection reagent (S1:inFluB / Victoria...
Embodiment 3
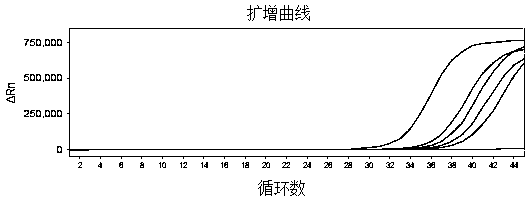
[0101] Example 3 specificity
[0102] The specificity was verified by using the primer-probe combination (Table 2), reagents and nucleic acid amplification detection method in Example 1.
[0103] In order to prove the cross-reactivity of the kit, human coronavirus OC43, human coronavirus 229E, human coronavirus NL63, human coronavirus HKU1, SARS coronavirus, MERS coronavirus, meningococcus, Haemophilus influenzae, gold Staphylococcus aureus, Streptococcus pneumoniae, Rubella virus, Mumps virus, Respiratory adenovirus type 3, Respiratory adenovirus type 7, Respiratory syncytial virus type B, Parainfluenza virus type 2, Streptococcus pyogenes, Bordetella pertussis, Candida albicans, Pseudomonas aeruginosa, Staphylococcus epidermidis and other 21 pathogens and human genome DNA were detected respectively.
[0104] Firstly, the virus was diluted to 1E+05 CFU / mL, and the bacterial and human genomic DNA was diluted to 1E+06 copies / mL, and then the nucleic acid of each pathogenic mic...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com