Primer probe composition for detecting methylation level of DNA (deoxyribonucleic acid) and application of primer probe composition
A primer-probe, composition technology, used in recombinant DNA technology, DNA/RNA fragments, determination/inspection of microorganisms, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Embodiment 1 is used to detect the design screening of the primer probe of METRNL, PDE4D and MAPT gene DNA methylation level
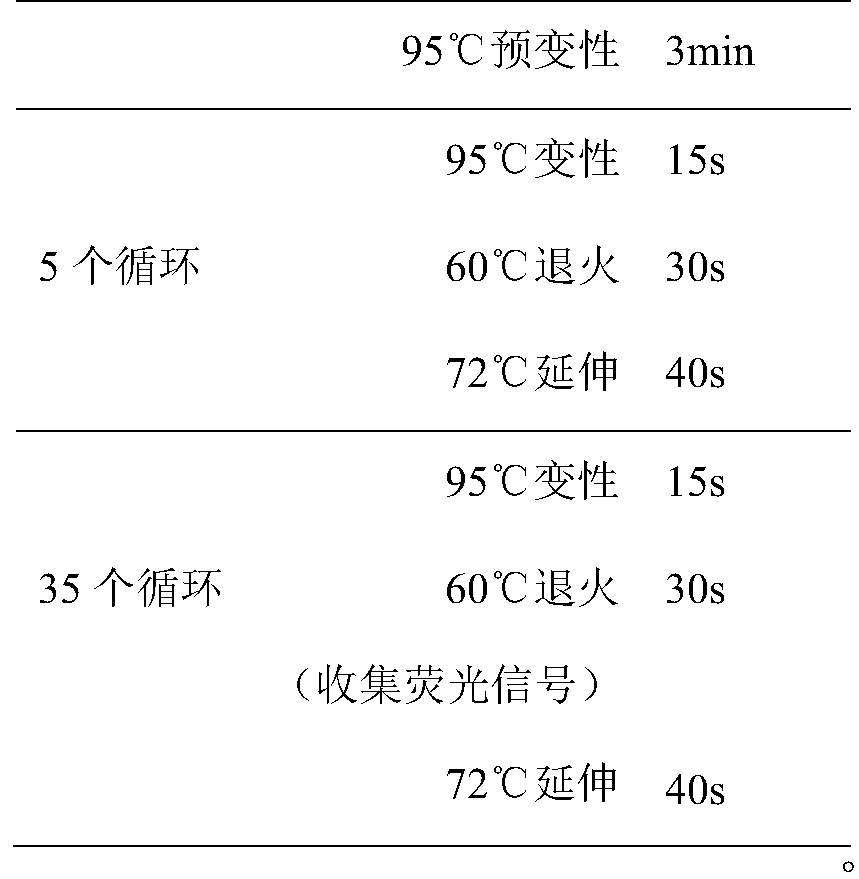
[0069] The main principle of the present invention is: in the reaction system of the fluorescent PCR-probe method, a pair of PCR amplification primers and a pair of probes with different fluorescent labels are included. The probe is only specifically combined with the template (ie sample DNA), and the probe binding region is between the primer pair. When the probe is intact, the fluorescent signal emitted by the 5'-end fluorescent group of the probe will be absorbed by the 3'-end quencher group, and the PCR instrument cannot detect the fluorescent signal. With the PCR amplification, the Taq enzyme will When encountering a probe that specifically binds to the template, the 3'-5' exonuclease activity of Taq enzyme will cut off the probe, and the fluorophore will be far away from the quencher group, so that the instrument can detect a specific fluo...
Embodiment 2
[0086] Embodiment 2 A kind of primer probe combination carries out the real-time fluorescent PCR detection method of METRNL, PDE4D and MAPT gene DNA methylation level
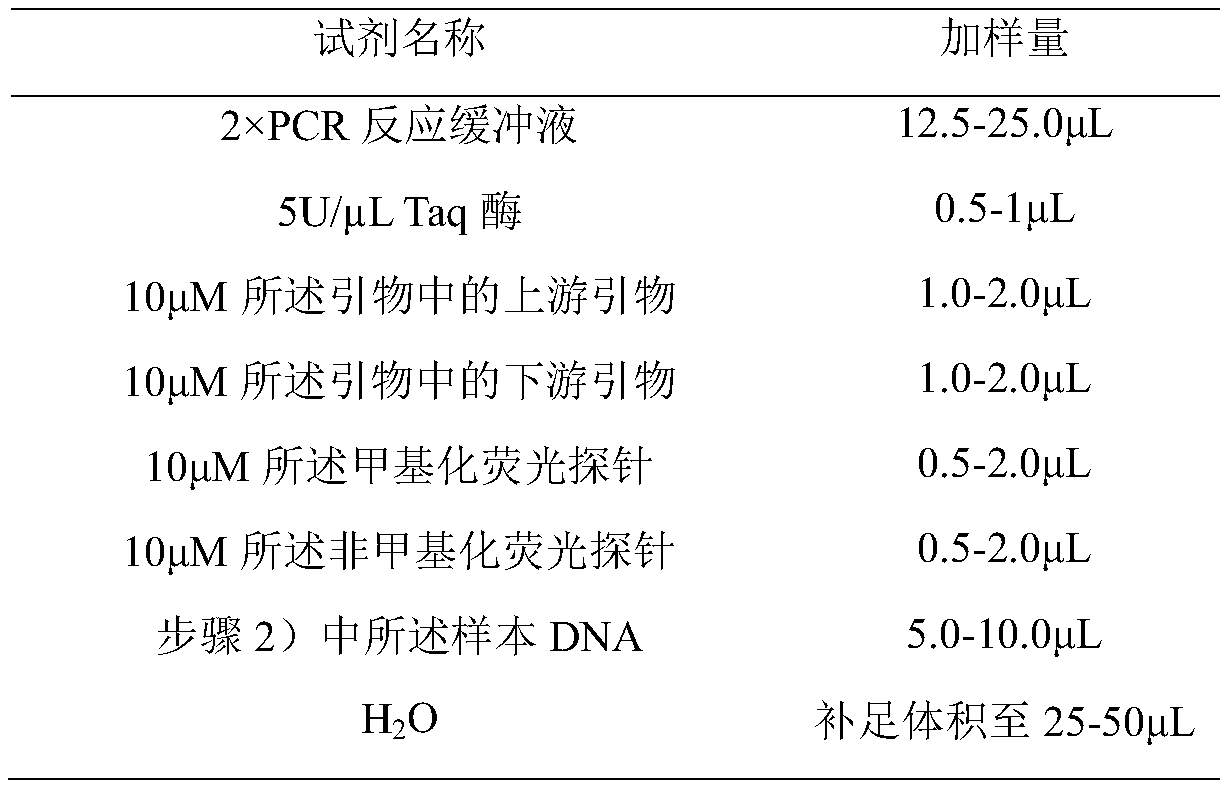
[0087] The method for selecting the primer probe combination described in Example 1 to detect the DNA methylation level of METRNL, PDE4D and MAPT genes comprises the following steps:
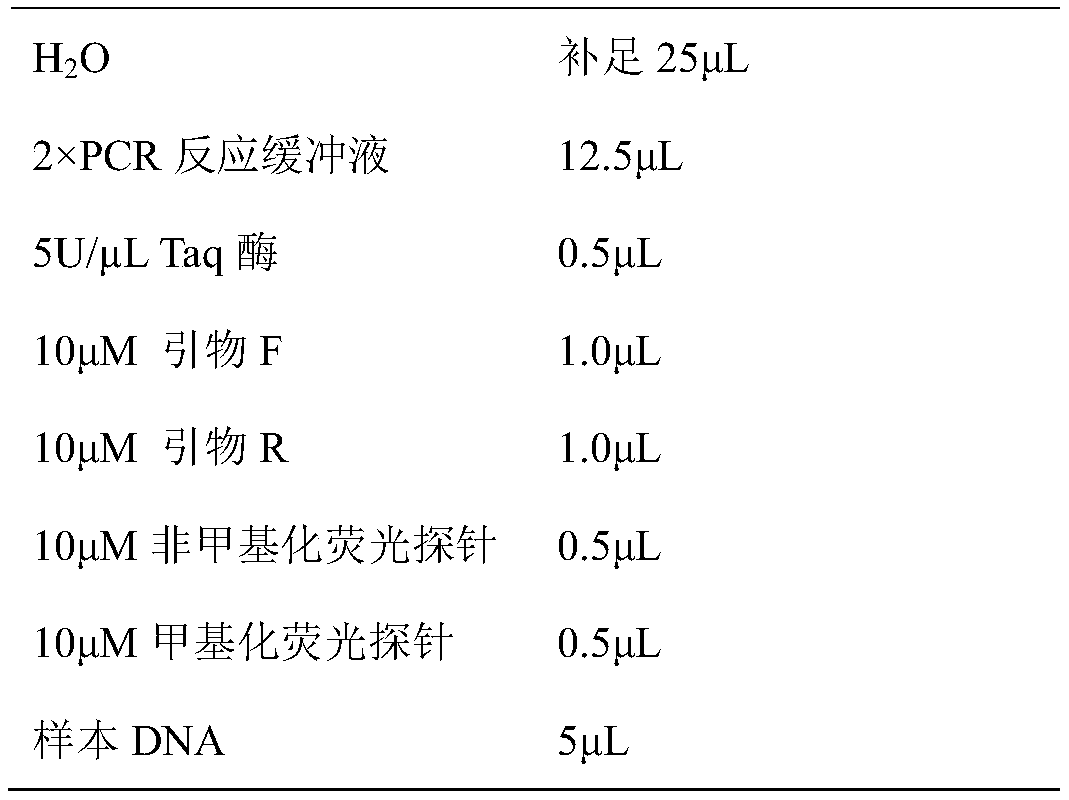
[0088] 1. Extract the genomic DNA in the sample: Use the blood genomic DNA extraction kit (0.1-1mL) (purchased from Tiangen Biochemical Technology (Beijing) Co., Ltd., model DP348) to extract the human genomic DNA in the human whole blood sample, and the genomic DNA is subjected to ultraviolet light After spectrophotometer detection, the concentration of genomic DNA is required to be not less than 40ng / μL, OD 260 / OD 280 The ratio is not less than 1.80;
[0089] 2. Sulfite modification of genomic DNA: Add 5.5 μL 3M NaOH solution to 50 μL genomic DNA sample, denature at 42°C for 30 minutes, add 30 μL 10 mM hydroquinone solution,...
experiment example
[0125] Experimental Example The effect of a combination of primers and probes on the detection of DNA methylation levels of METRNL, PDE4D and MAPT genes
[0126] According to the detection method provided in Example 2, 3 different samples were tested, and the sources of samples 1-3 were different peripheral blood DNAs clinically obtained by Xuanwu Hospital.
[0127] The detection results of the methylation level at positions 1-4 of the CpG island of each gene in each sample are as follows:
[0128] Sample 1(%) Sample 2(%) Sample 3(%) METRNL gene CpG island position 1 28 28 29 METRNL gene CpG island position 2 27 25 27 METRNL gene CpG island position 3 29 27 20 PDE4D gene CpG island position 1 33 31 31 PDE4D gene CpG island position 2 40 38 39 PDE4D gene CpG island position 3 26 31 29 PDE4D gene CpG island position 4 34 36 36 MAPT gene CpG island position 1 94 94 94 MAPT gene CpG island position ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com