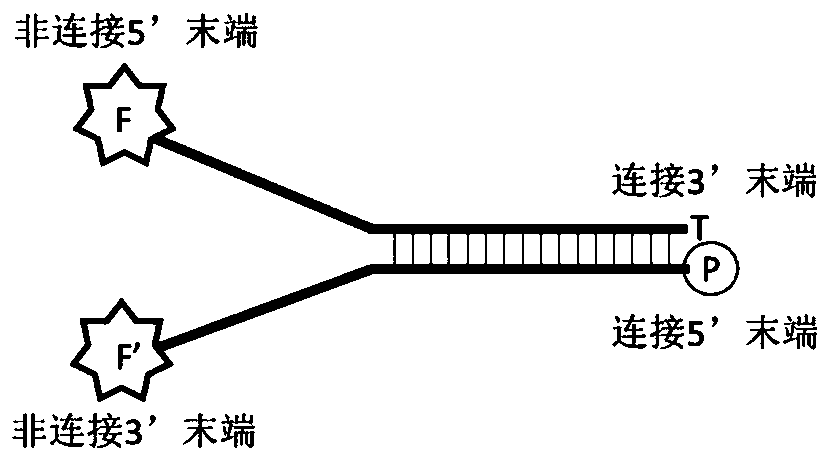
DNA (deoxyribonucleic acid) connector as well as preparation method and application thereof
A DNA sequencing and construction method technology, applied in the fields of DNA adapters and their preparation and application, can solve problems such as unusability and complexity, and achieve the effects of avoiding cost, simple construction method and improving connection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
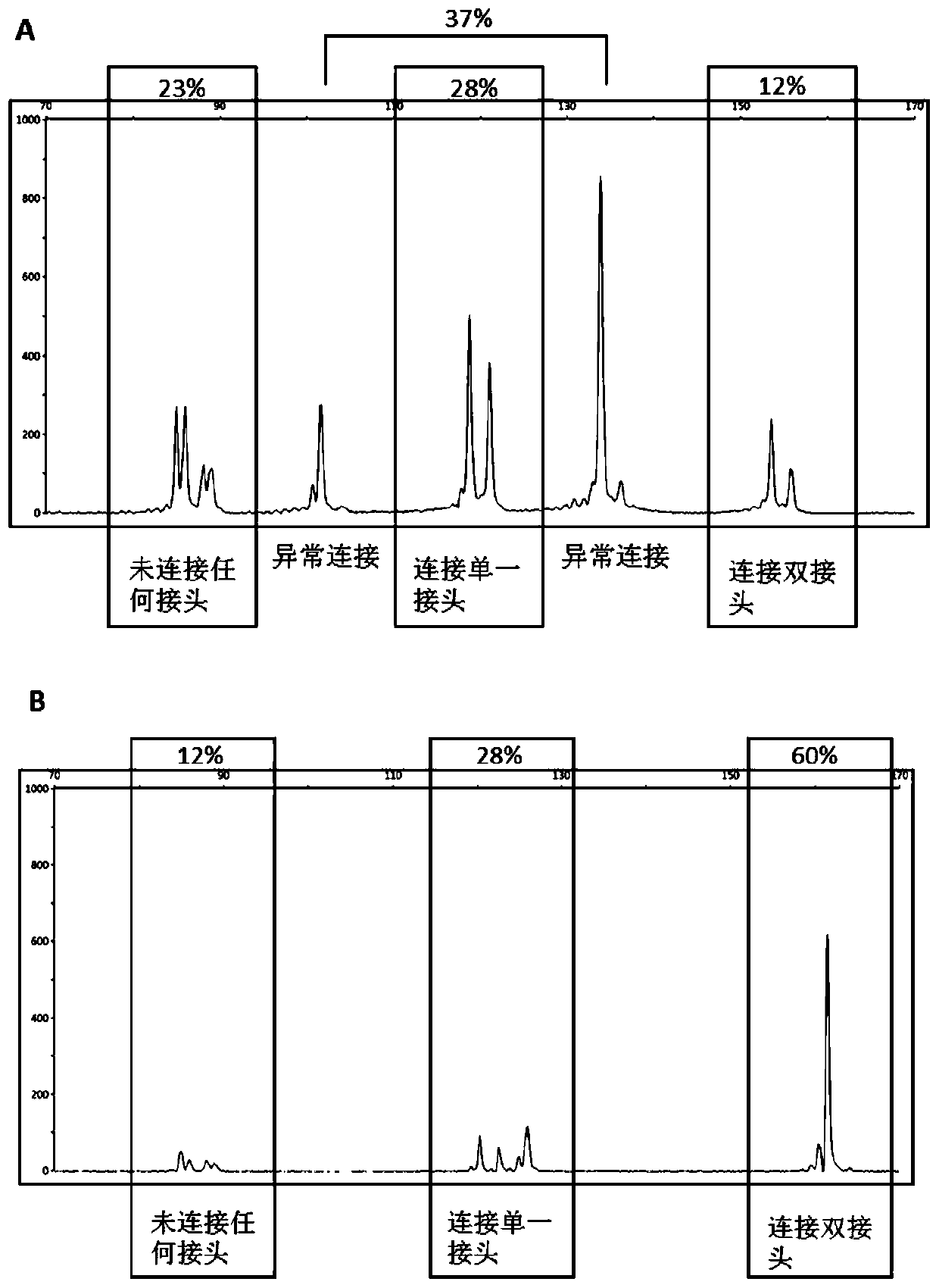
[0036] Example 1: Capillary electrophoresis verification of DNA connection efficiency of conventional Y-shaped adapters
[0037] 1. Synthesize the following primer sequences
[0038] DNAQC-F: ccg GAATTC TT[6-FAM-dT]GCCTTCATTGAGCGCTACTT (SEQ ID NO.1)
[0039] DNAQC-R: aaaa CTGCAGTTCCAGGGTCTTTCTCAATCCAG (SEQ ID NO. 2)
[0040] 2. Using the Thermus aquaticus genomic DNA as a template, PCR amplification was carried out with the above primers, and after amplification, a specific double-stranded DNA with a length of 89 bp was obtained.
[0041] 3. After purification using DNA Clean&Concentrator-5 (200Preps) w / Zymo-Spin ICColumns (Capped) from Zymo Company, the quality control DNA fragment obtained was named DNAQC.
[0042] 4. The following linker sequences were chemically synthesized and purified by HPLC.
[0043] Basic-F: ACACTCTTTCCCTACACGACGCTCTTCCGATCT (SEQ ID NO. 4)
[0044] Basic-R: GATCGGAAGAGCACACGTCTGAACTCCAGTCAC (SEQ ID NO. 5)
[0045] Basic-F-F: [ROX] ACACTCTTTCCCTAC...
Embodiment 2
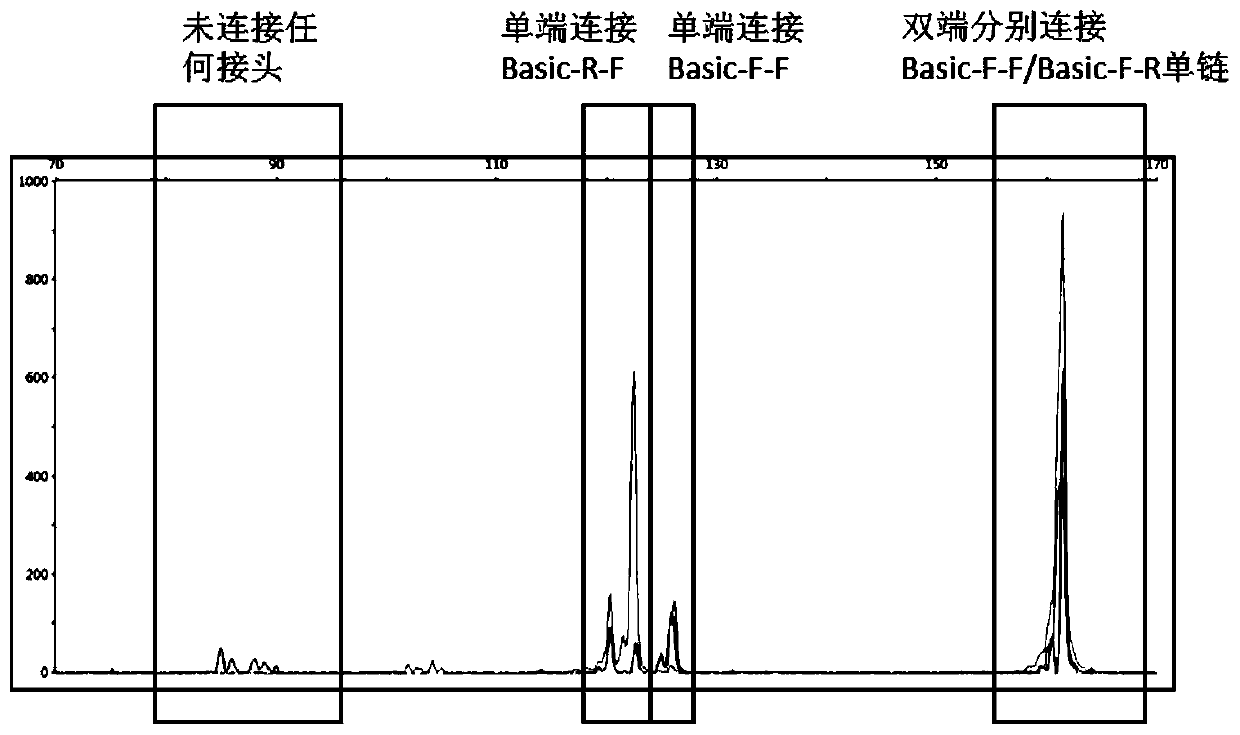
[0064] Example 2: Capillary Electrophoresis Verification of DNA Ligation Efficiency Containing UMI Sequence Y-shaped Adapter
[0065] 1. Prepare DNA QC with Example 1.
[0066] 2. Chemically synthesize the following fragments and purify using HPLC
[0067] DS-F: [ROX] AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT (SEQ ID NO. 8)
[0068] DS-R: TCTTCTACAGTCANNNNNNNNNNNNAGATCGGAAGAGCACACGTCTGAACTCCAGTCAC [HEX] (SEQ ID NO. 9)
[0069] 3. Use H 2 O Dissolve the DS-F / DS-R synthesized above to a final concentration of 100 μM. Take 25 μl of DS-F and 25 μl of DS-R and mix them into 50 μl of DS-AD (final concentration 50 μM) in a 0.2 ml thin-walled tube.
[0070] 4. Place the DS-AD above in a PCR machine for annealing, the condition is denaturation at 95°C for 10 minutes, cooling down to 25°C at 0.1°C / sec; and keeping at 25°C for 2 hours.
[0071] 5. Prepare the system described in Table 5 below for single-strand extension
[0072] table 5
[0073] 50μl A...
Embodiment 3c
[0082] Example 3 cfDNA low-input library construction
[0083] 1. Use the MagMAX cell-free DNA isolation kit from Thermo Fisher to extract cfDNA from 5ml plasma.
[0084] 2. Referring to Example 1, Synthesize Basic-AD and Basic-AD-F.
[0085] 3. Take 5ng and 1ng cfDNA respectively, use conventional library construction reagents for library construction, and prepare the following system in Table 7 to fill in the ends and add A
[0086] Table 7
[0087] 5μl Terminal balance + A buffer 1μl T4 DNA polymerase 1μl T4 PNK 1μl Taq 1ng / 5ng cfDNA Make up to 30μl h 2 o
[0088] Reaction conditions: 20°C for 30 minutes, 70°C for 30 minutes.
[0089] 4. After the end is filled and A is added, the connector is connected, and H is used first 2 O Dilute Basic-AD and Basic-AD-F to 2 μM, and prepare the following system in Table 8:
[0090] Table 8
[0091] 30μl End filling + A segment 26μl ligation buffer 3μl ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com