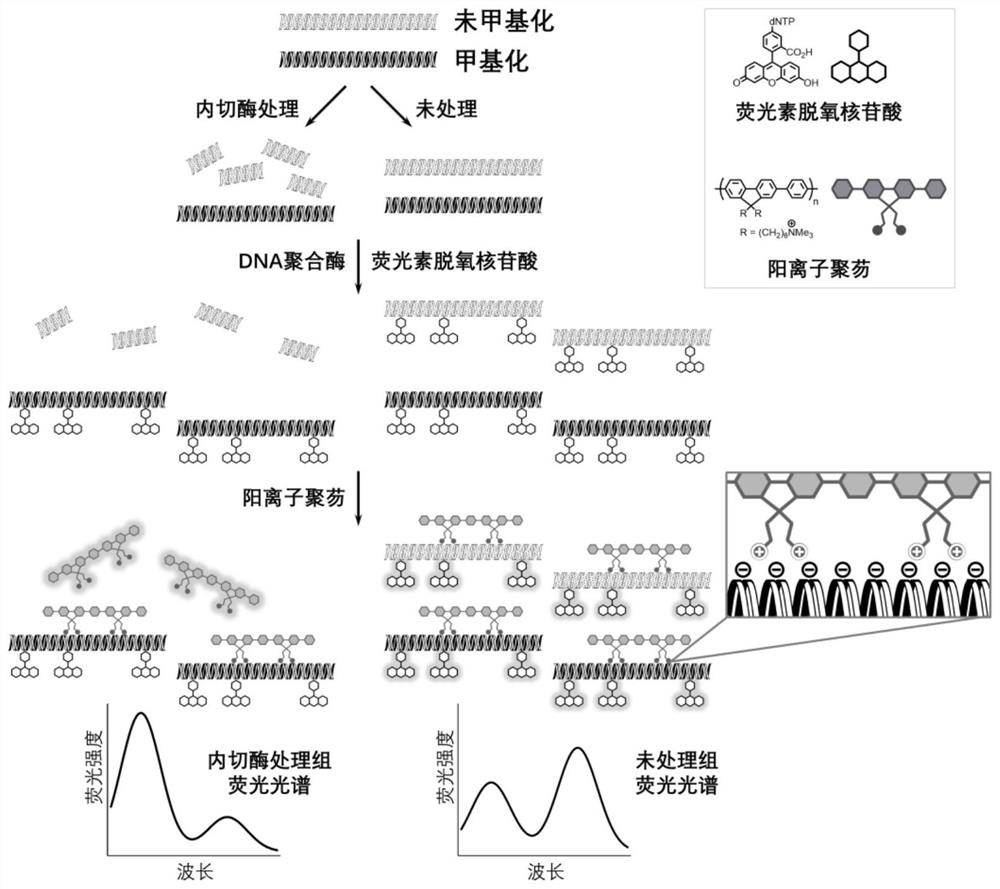
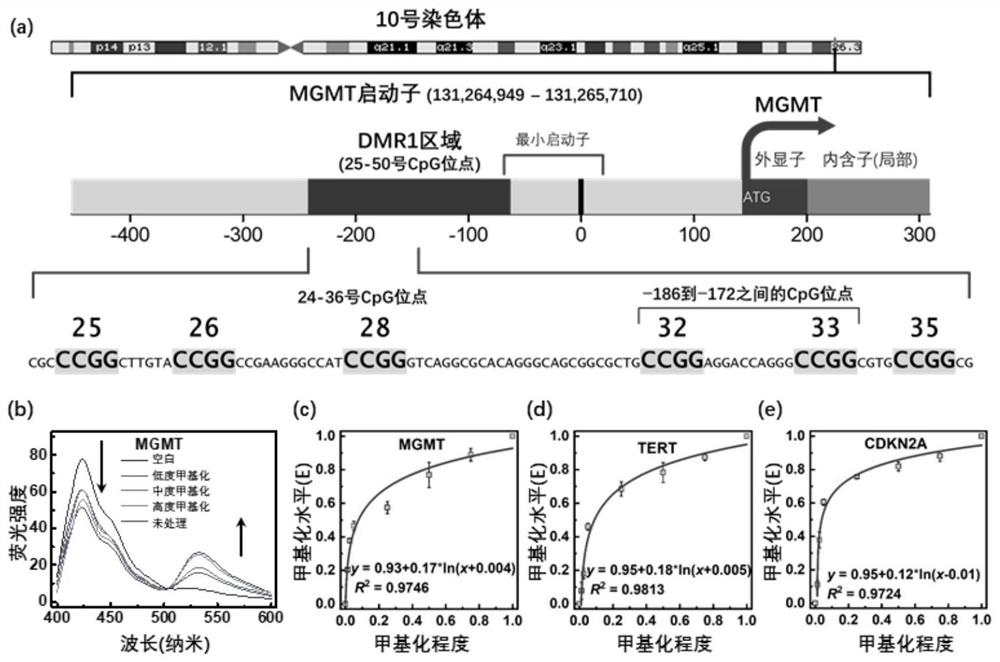
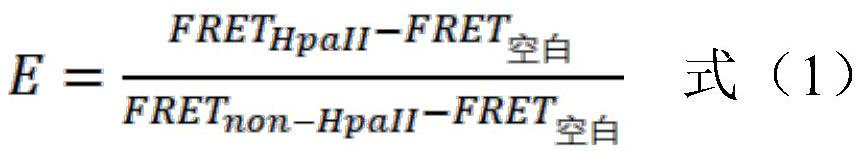Method for detecting gene methylation in glioma
A technology of glioma and methylation, applied in the direction of biochemical equipment and methods, microbial measurement/testing, etc., can solve the problems of insufficient sensitivity, low cost, high cost, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] 1. Collection of specimens, DNA extraction and preparation
[0060] ①Sample collection and storage: After the glioma sample was removed, it was immediately stored in pieces in a -80°C refrigerator for later use.
[0061] ② DNA extraction
[0062] Take 25 mg of specimen tissue, cut it into pieces and process it into a cell suspension, put it into a 1.5 ml centrifuge tube; add 180 μl of ATL buffer, shake until completely suspended.
[0063] Add 20 μl proteinase K solution with a concentration of 20mg / μl, mix well, incubate at 56°C until the tissue is completely dissolved, add 4 μl RNase A, and mix well.
[0064] Add 200 μl of AL buffer and alcohol (about 98%), and mix well.
[0065] Add all the solution obtained in the previous step to the DNease adsorption column, add 500 μl AW1 buffer, centrifuge at 12000 rpm for 1 min, discard the waste liquid, and put the adsorption column back into the collection tube.
[0066] Add 500 μl AW2 buffer solution to the DNease adsorpti...
Embodiment 2
[0106] According to the method of Example 1, the difference is that the weight ratio of the PCR product to the cationic conjugated polymer in step (3) is 1:7.68. And record the lowest DNA concentration and the lowest methylation level that it can detect.
[0107] The results are shown in Table 7.
[0108] The result of table 7 embodiment 1
[0109]
[0110]
[0111] *The TERT methylation level of the sample cannot be detected within the detection range of this method, so it is expressed as 0%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com