Novel coronavirus variation analysis method and application
A coronavirus and mutation analysis technology, applied in the field of novel coronavirus mutation analysis, can solve the problems of high error rate of mutation detection, lack of new coronavirus mutation detection software, third-party general software cannot directly process metagenomic sequencing data, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
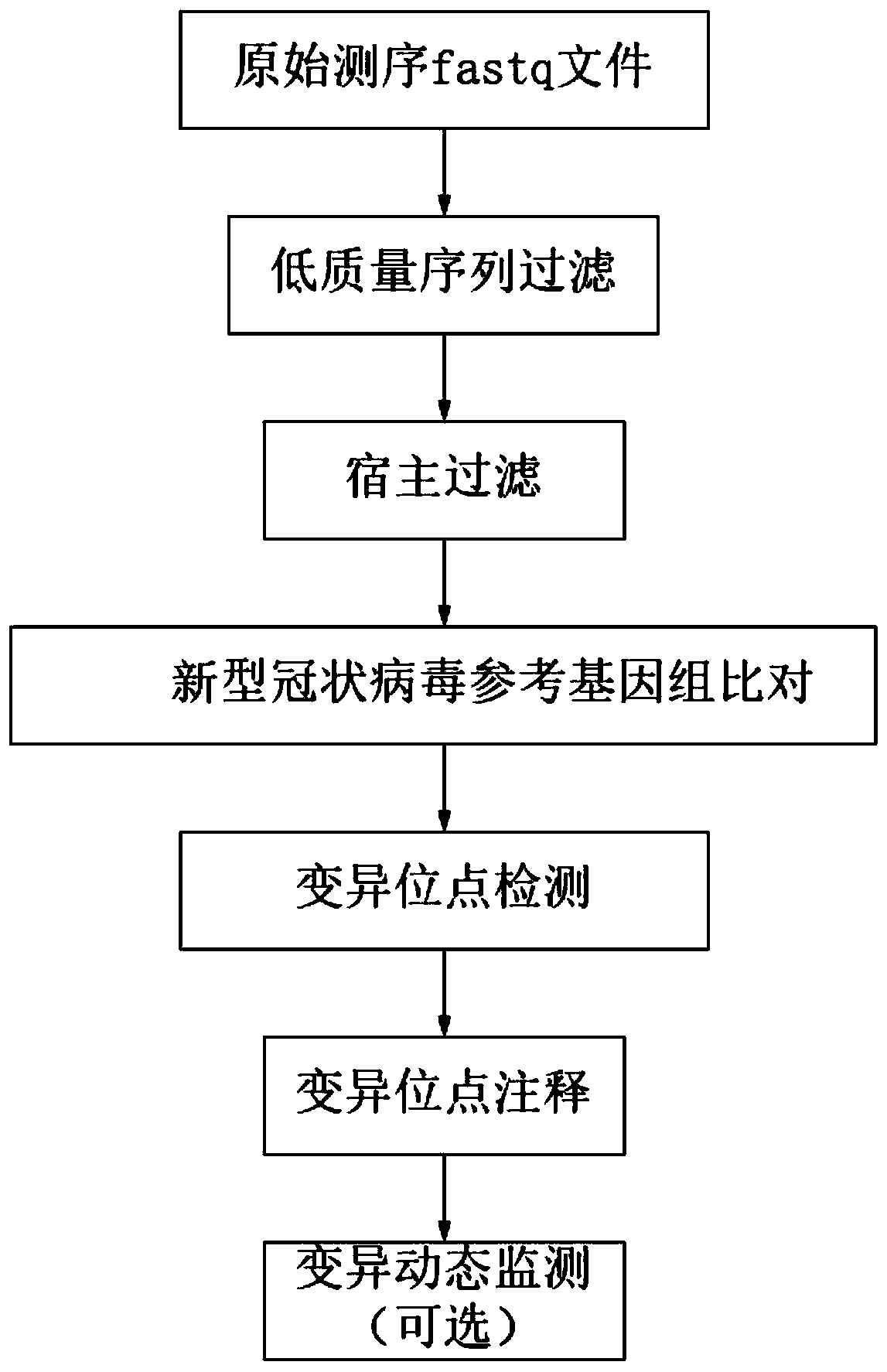
[0060] A novel coronavirus (2019-nCoV) variation analysis method, comprising the following steps:
[0061] 1. Database construction
[0062] 1. Host database construction
[0063] Download the human reference genome (accession number: GCF_000001405.39) from the NCBI website (https: / / ftp.ncbi.nlm.nih.gov / genomes), and use the bwa index software to construct an index file to obtain the host database.
[0064] 2. Construction of the Novel Coronavirus Reference Genome Database
[0065] Download the new coronavirus (2019-nCoV) reference genome from the NCBI website, the accession number is GCF_009858895.2, and download the corresponding annotation file in gtf format. Use the bwa index to construct a novel coronavirus reference genome database.
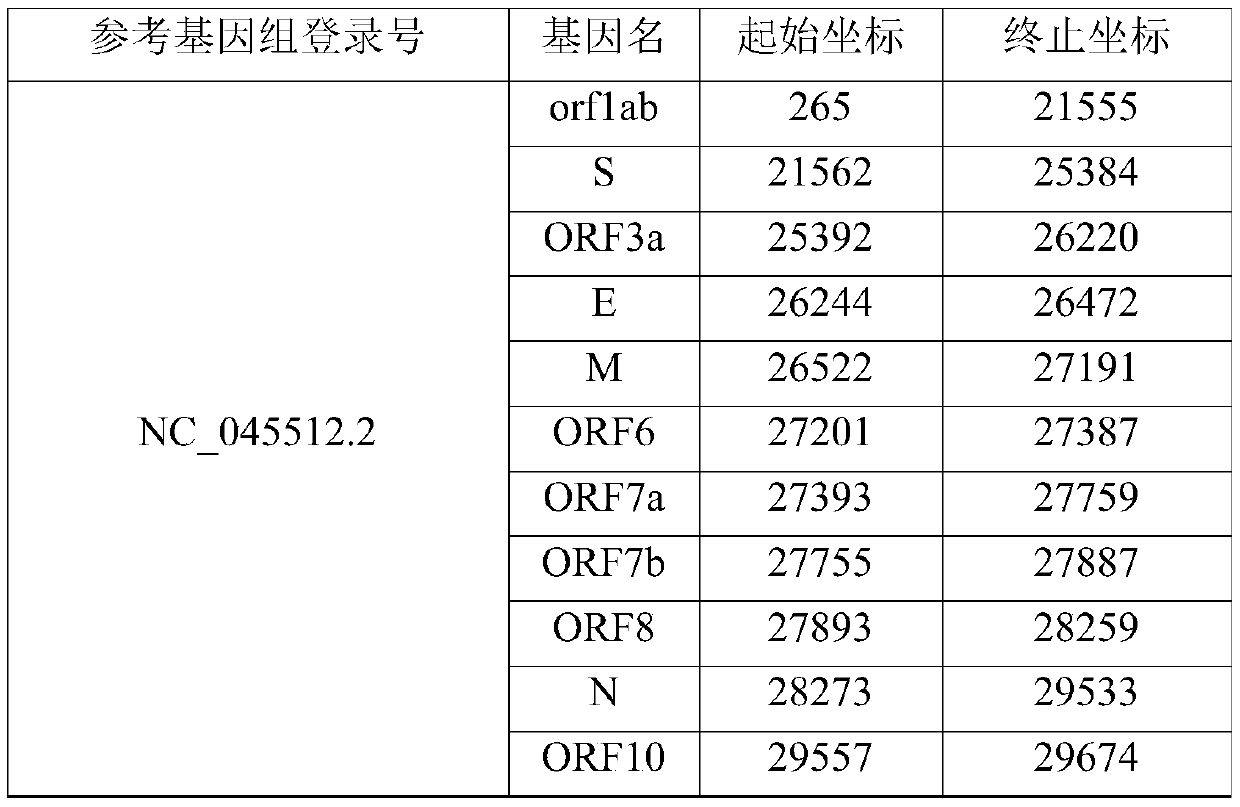
[0066] The gene coordinates of this new coronavirus reference genome are shown in the table below.
[0067] Table 1. Gene coordinate information of the new coronavirus
[0068]
[0069] Extract the gtf file gene coordinates, coding ...
Embodiment 2
[0139] Evaluation of the accuracy of novel coronavirus mutation detection.
[0140] The combination of GATK and snpEff software is the most commonly used method with high accuracy in the field of microbial variation detection and annotation, and is called a classic method by the industry.
[0141] In order to evaluate the accuracy of the method of the present invention in detecting the mutation of the new coronavirus, it is most reasonable to analyze the virus with known mutation information. Therefore, this example will evaluate the present invention by comparing the variation detection results of the classical method and the method of the present invention accuracy of the method.
[0142] The above-mentioned classical method and the method of the present invention are used to analyze the sequencing data of the same known virus variation information.
[0143] The mutation information of the virus is shown in Table 6 below. There are 8 mutation sites in total, all of which ar...
Embodiment 3
[0153] Dynamic monitoring of novel coronavirus mutations.
[0154] This example will use simulated data to illustrate the application of the present invention in the dynamic monitoring of novel coronavirus mutations.
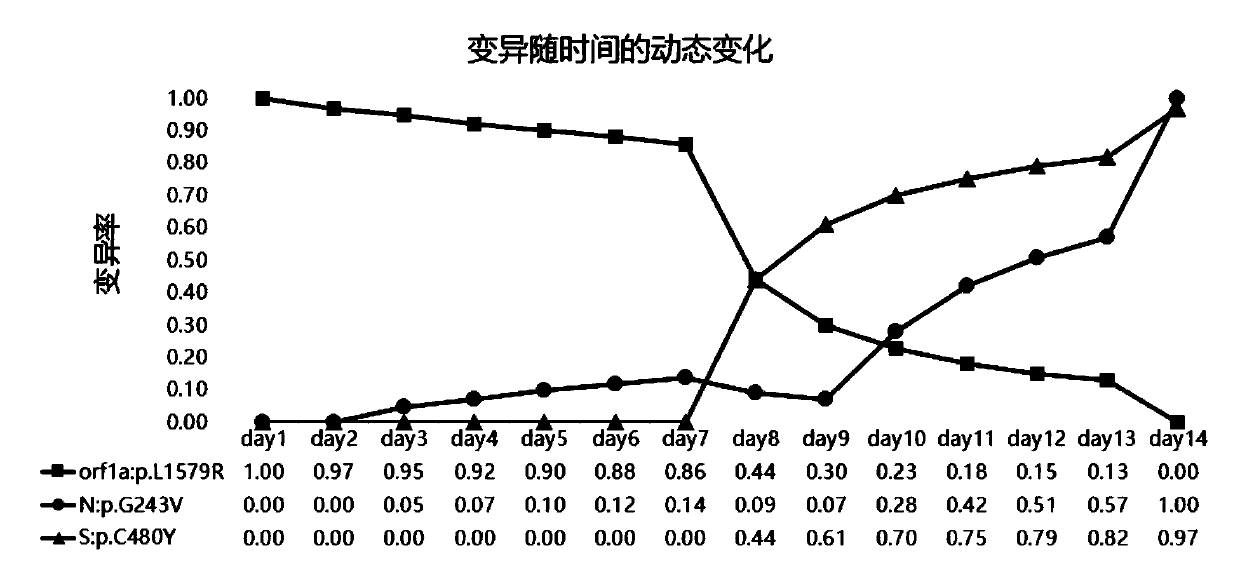
[0155] The duration of a patient infected with the new coronavirus is generally 14 days. A simulated continuous sampling of a patient with new coronary pneumonia for 14 days obtained 14 samples, which were named day1~day14 according to the order of time collection, and metagenomic sequencing was performed to detect the mutation of the new coronavirus. The method of Example 1 analyzes the dynamic process of variation.
[0156] Test results such as figure 2 shown. The results showed that 3 mutations were detected in 14 consecutive samples, namely orf1a:p.L1579R, N:p.G243V, and S:p.C480Y.
[0157] On day1 and day2, there was only one mutation, orf1a:p.L1579R, and the ratio was close to 1. A new mutation N:p.G243V appeared from day3, and a new mutation S:p.C480Y a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com