Gene marker for monitoring water pollution source in real time and application of gene marker
A genetic marker and real-time monitoring technology, applied in the field of bio-environmental technology detection, can solve the problems of being unable to locate water pollution sources and monitor water pollution in real time, and achieve excellent sensitivity effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1 Primer Design
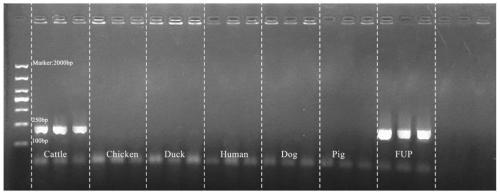
[0029] 224 animal feces samples (chicken, duck, human, dog, pig, cow) were collected, and 984,224 bacterial 16SrDNA sequences were obtained by high-throughput sequencing. By comparing the obtained sequences to the database, the 16SrDNA gene sequences of Faecalibacterium in all feces samples were selected, and a detailed comparison between species was performed, and a pair of DNA primers Cattle-1 (amplification length: 214bp) were designed to specifically identify cow feces. ), design the forward primer as shown in SEQ ID NO:1, and the reverse primer as shown in SEQ ID NO:2.
Embodiment 2
[0030] Embodiment 2 monitors pollution source
[0031] 1) DNA extraction:
[0032] DNA was extracted from 224 stool samples of 6 species (MO-BIO DNA Isolation kit);
[0033] 2) Specificity verification:
[0034] Get the genomic DNA in step 1), mix the DNA samples of the same species, respectively Cattle-1, FUP (universal primer, forward primer as shown in SEQ ID NO:3, reverse primer as shown in SEQ ID NO:4 Shown) as primers, add 7.5 microliters of TaqPCR Master Mix (2×), 0.5 microliters of forward primers, 0.5 microliters of reverse primers, and 1 microliters of genomic DNA; and add sterile water to 15 microliters; PCR reaction , the reaction temperature is as follows
[0035] Cattle-1: Pre-denaturation at 96°C / 300s, 1 cycle; denaturation at 96°C / 30s, annealing at 56°C / 30s, 32 cycles.
[0036] Such as figure 1 As shown in the primer specificity diagram, primer Cattle-1 reacts with Faecalibacterium in cow feces to produce bands, and there is no band in other species, in...
Embodiment 3
[0037] Example 3 Positive Rate Determination
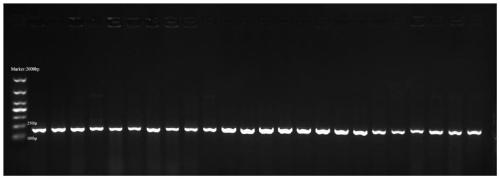
[0038] DNA was extracted from 48 cow feces samples for detection, such as image 3 Among the 48 DNA samples shown, 44 samples can be PCR amplified by primer Cattle-1 and obvious bands can be seen, and the positive rate is 91.7%, which shows that the primer Cattle-1 has good stability.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com