A polypeptide specifically binding to Mycobacterium tuberculosis, its encoding gene and application
A Mycobacterium tuberculosis, coding gene technology, applied in the field of biomedicine, can solve problems such as unsatisfactory diagnosis and analysis, and achieve the effect of low cost, simple preparation process and high stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1 Screening of polypeptides that bind to MA
[0055] 1. Amplification and purification of phage library
[0056] inoculation E. coli ER2738 single colonies were incubated in 5-10mL LB liquid medium, 37°C, 200rpm shaker to mid-logarithmic phase (OD 600 ≈0.5); add 10 μL of phage, shake at 37°C and 200rpm for 4-5h, centrifuge at 10,000g for 10min, and take the supernatant; centrifuge again at 10,000g for 10min, take 80% of the supernatant, add 1 / 6 volume of PEG8000 / NaCl, 4°C Let stand overnight. Take it out the next day, and the white precipitate is the phage. Centrifuge at 10000g for 15 minutes, discard the supernatant, and gently suck out the residual solution after brief centrifugation; add 1mL TBS solution to dissolve the white precipitate.
[0057] 2. Phage Titer Determination
[0058] inoculation E. coli ER2738 single colonies were incubated in 5-10mL LB liquid medium at 37°C and 250rpm on a shaker until mid-log phase (OD 600 ≈0.5); Microwave heati...
Embodiment 2
[0065] Example 2 Identification of MA-specific phage
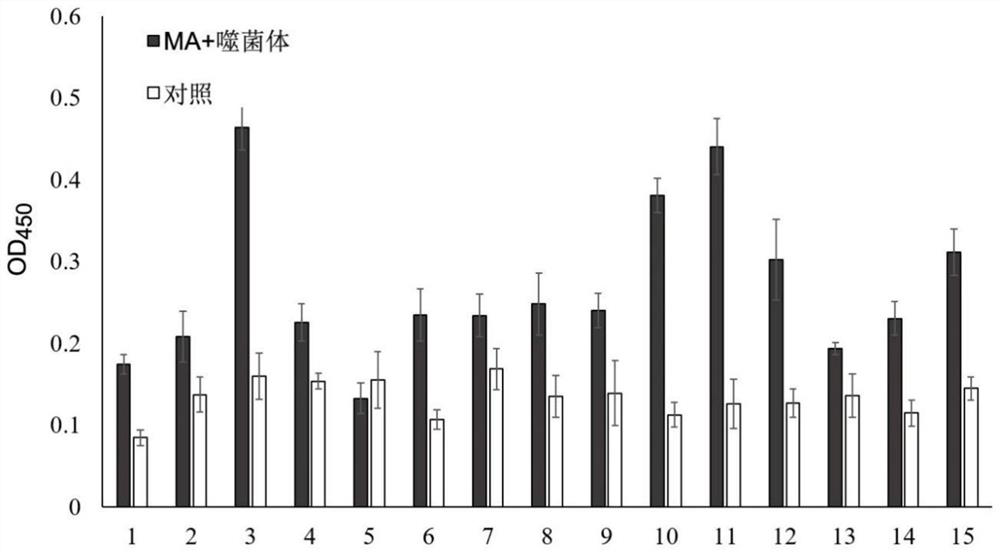
[0066] Randomly pick 20 well-separated single colonies on the titer determination plate after the third round of screening in Example 1, and after culturing and purifying separately, use phage ELISA to detect the binding activity of each strain of phage to MA. The specific steps are as follows:
[0067] Coat the ELISA plates with MA and Blocking respectively, and each group was paralleled three times, and blocked overnight at 4°C, washed 6 times with TBST, added 100 μL of purified phage, incubated at 37°C for 1 hour with shaking, washed 10 times with TBST to remove unbound For phage, add 100 μL of HRP-labeled mouse anti-M13 monoclonal antibody, incubate with shaking at 37°C for 1 hour, wash 10 times with TBST, add TMB substrate and react in the dark at room temperature for 5-10 minutes, stop the reaction with 2mol / L sulfuric acid, and measure the OD value at a wavelength of 450nm , with P / N ≥ 2.1 as positive.
[0068] Te...
Embodiment 3
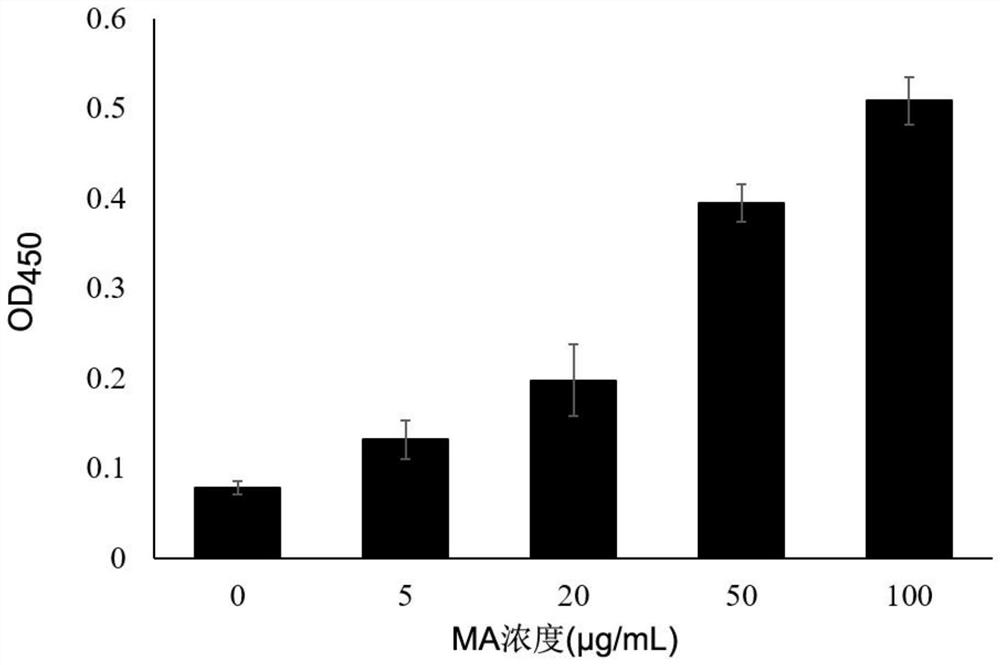
[0073] Example 3 Determination of binding activity of polypeptide sequences
[0074] The plates were coated with different concentrations of MA and Blocking and blocked overnight at 4°C, washed 6 times with TBST, and 2×10 phages with the Thanos1 sequence were added. 11 (with the same titer of phage without Thanos1 sequence as the control), wash 10 times with TBST to remove unbound phage, add 100 μL of HRP-labeled mouse anti-M13 monoclonal antibody, incubate with shaking at 37°C for 1 hour, wash 10 times with TBST; TMB was reacted at room temperature in the dark for 5-10 minutes, 2mol / L sulfuric acid was used to terminate the reaction, and the OD value was measured at a wavelength of 450nm. The result is as figure 2 As shown, phages with the Thanos1 sequence exhibited specific binding to MA.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com