Halohydrin dehalogenase mutant and application thereof in synthesis of chiral epichlorohydrin
A technology of halohydrin dehalogenase and mutants, which is applied in the application field of preparing epichlorohydrin, can solve the problems such as low e.e. value of halohydrin dehalogenase catalysis, product racemization, etc., and achieve industrial production, Enzyme activity improvement, the effect of enzyme activity improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1: Homology Modeling and Molecular Dynamics Simulation of Halohydrin Dehalogenases
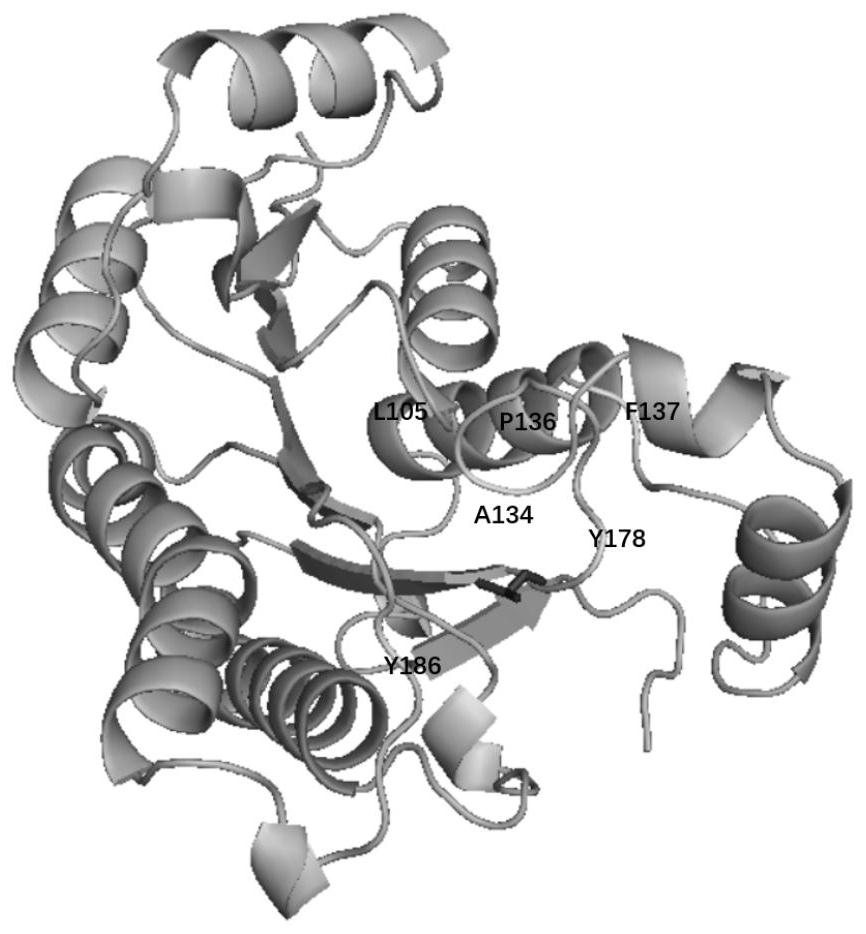
[0030] Using the three-dimensional structure of HheC (PDB ID: 1ZO8) derived from Agrobacterium radiobacterstrain AD1 in GenBank AAK92099 as a template, the three-dimensional structure of 1,3-dcp was drawn using ChemDraw 8.1, and the molecular docking program was aligned according to the AutoDock 4.6.2 program. Docking of HheC and substrate 1,3-dcp. All the above structures were visually analyzed using the PyMOL program, and it was found that the 105th, 134th, 136th, 137th, 178th, and 186th positions are the key sites that affect the enzyme activity ( figure 1 ).
Embodiment 2
[0031] Example 2: Construction of Halohydrin Dehalogenase Site-Directed Saturation Library
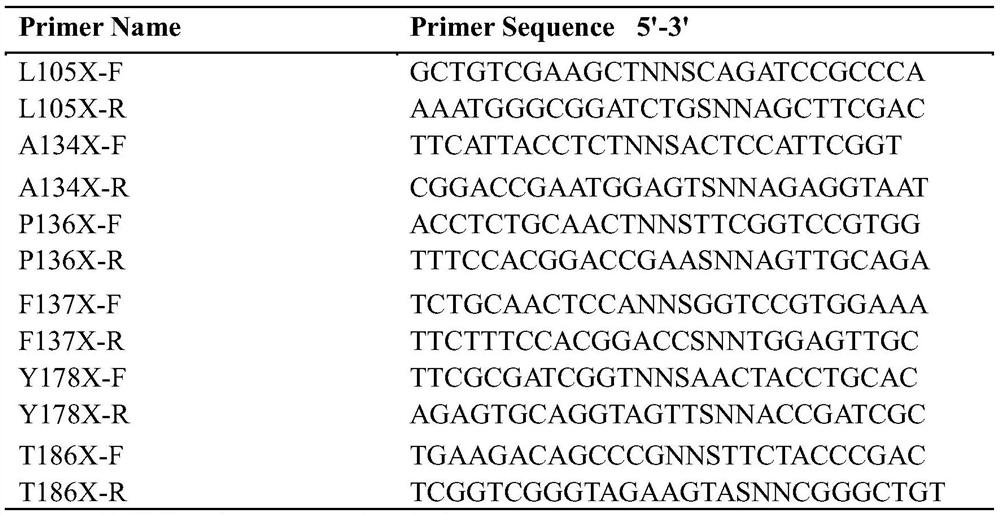
[0032]According to the conclusion of Example 1, design based on the gene sequence (nucleotide sequence shown in SEQ ID NO.1, amino acid sequence shown in SEQ ID NO.2) of the wild-type halohydrin dehalogenase HheC included in GenBank Primers (see Table 1). Respectively with primers L105X-F / L105X-R, A134X-F / A134X-R, P136X-F / P136X-R, F137X-F / F137X-R, Y178X-F / Y178X-R, T186X-F / T186X-R The parental HheC gene (nucleotide sequence is SEQ ID NO.1) was subjected to site-directed saturation mutation, and pET-28b(+) was used as the expression vector to obtain mutant plasmids with the target gene, and the target gene The mutant plasmid was transformed into E.coli BL21(DE3), and the mutants of recombinant bacteria containing the halohydrin dehalogenase mutant gene were respectively obtained, which were respectively E.coli BL21(DE3)-L105X (referred to as mutant L105X), E. .coli BL21(DE3)-A134X (ref...
Embodiment 3
[0047] Example 3: Screening of Halohydrin Dehalogenase Mutant Library
[0048] 1. Screening of the halohydrin dehalogenase mutant library Taking wild-type HheC before mutation as a reference, pick a single colony clone (the mutant library constructed in Example 2) and culture it in a 2mL deep 96-well plate. 600 μL of LB culture solution with a concentration of 50 μg / mL kanamycin, and at the same time pick two parental strains in the last two wells of a 96-well plate as controls. Place a 2 mL 96-well plate at 37°C for 8 hours as a seed solution, then add 200 μL of the seed solution to a new sterile 600 μL LB culture solution containing a final concentration of 50 μg / mL kanamycin and 0.1 mM IPTG , after induction of expression at 28°C containing the seed solution for 12 hours, centrifuge at 4000 rpm for 20 minutes, discard the supernatant, collect the wet cells, and carry out the next step of high-throughput screening.
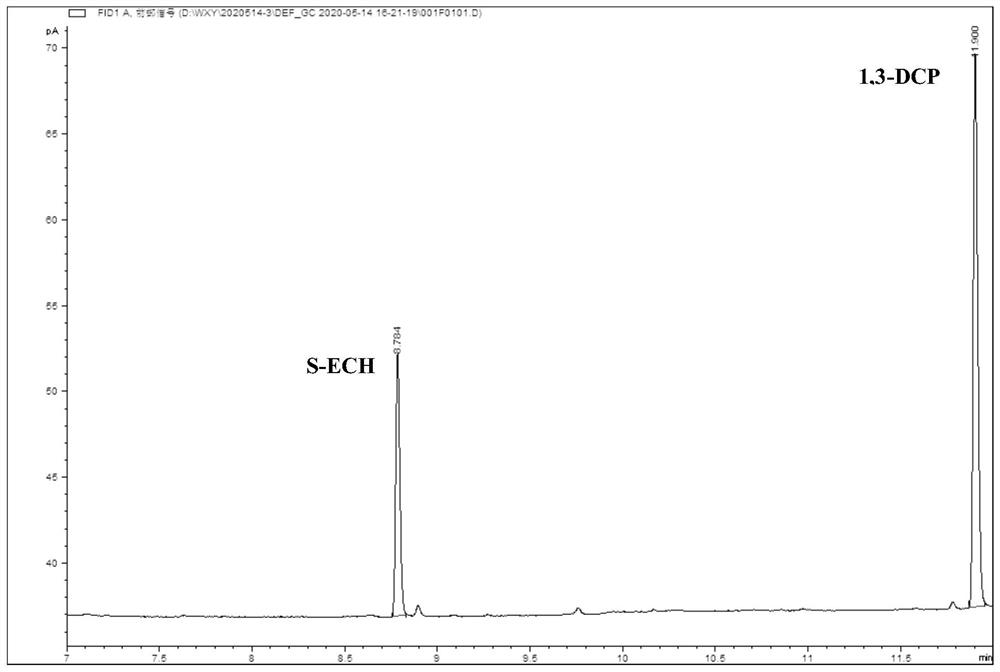
[0049] 2. According to the characteristics of the catalyt...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Upstream primer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com