High-throughput sequencing database construction technology capable of efficiently capturing three-dimensional conformation of chromosomes of frozen biological sample
A biological sample, high-throughput technology, used in the determination/inspection of microorganisms, biochemical equipment and methods, chemical libraries, etc., can solve the problem of 3D genome research with fewer frozen samples, achieve convenient collection and preservation, and improve library data. The effect of increasing quality and storage capacity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Example 1, Hi-C high-throughput sequencing of cotton
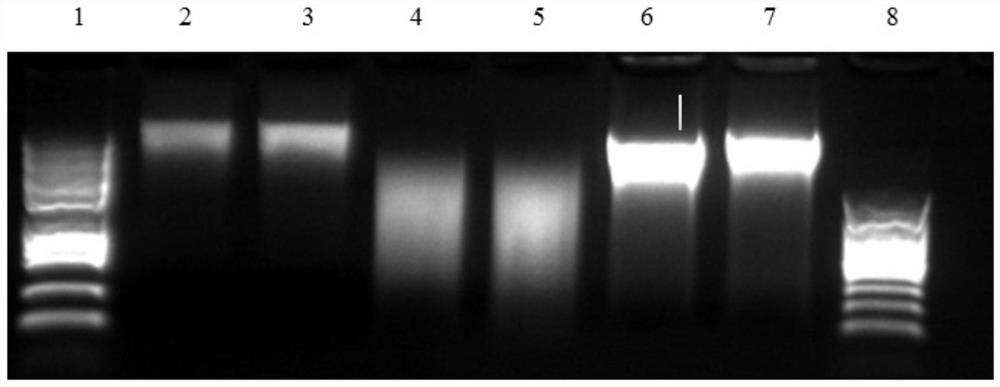
[0072] In this embodiment, the callus in the cotton tissue culture process was taken as the research object, and two batches of experiments were carried out, wherein the first batch of Hi-C libraries of the two methods was sequenced on a small scale (10g data volume, each Two small data evaluations) to evaluate the quality of the library, and found that the cotton Hi-C library of the new Hi-C method is much higher in the effective data ratio than the traditional method; in the second batch, we built two methods for cotton The libraries (three each) were deeply sequenced, and the genome coverage was approximately 75X, 50X and 30X (FS-Hi-C); 50X, 50X and 30X (traditional Hi-C). It was found that FS-Hi-C significantly increased the effective data ratio of the Hi-C library, and greatly reduced the ratio of repetitive sequences caused by PCR amplification in the library.
[0073] 1. The first batch of tests
[0074] (1...
Embodiment 2
[0216] Example 2, Hi-C high-throughput sequencing of Drosophila
[0217] In this example, Drosophila was used as the research object.
[0218] 1. Sample pretreatment
[0219] The method of the invention :
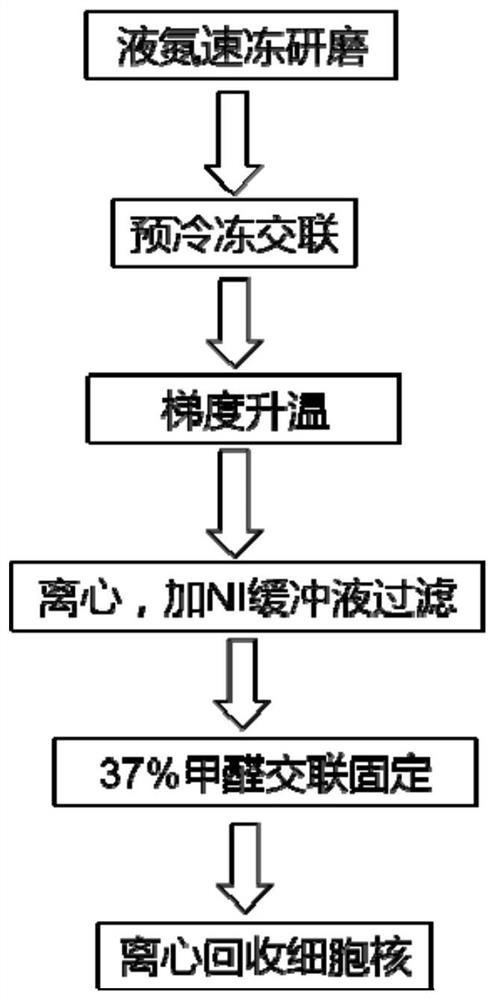
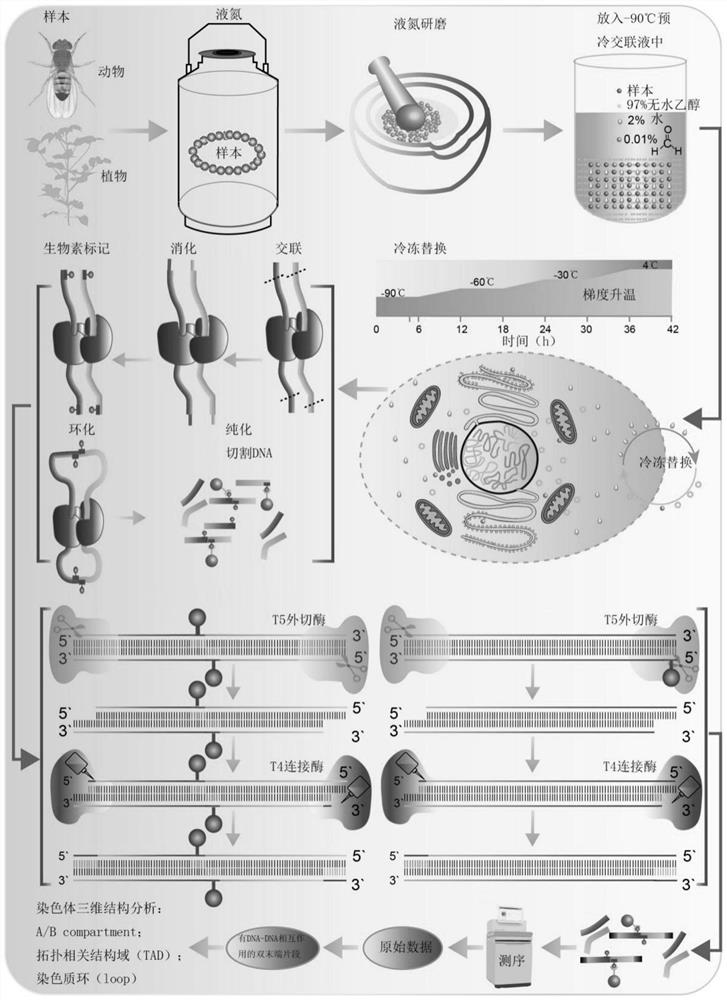
[0220] 1. Take 5×10 Drosophila cell line (S2 cell line) 6 cells were frozen in liquid nitrogen and ground into a powder.
[0221] 2. Transfer the grinding material to the pre-frozen cross-linking solution at -90°C, and then slowly increase the temperature: -90°C, 6h; -90°C to -60°C, 6h (5°C per hour); -60°C , 6h; -60°C to -30°C, 6h (5°C per hour); -30°C, 6h; -30°C to 0°C, 6h (5°C per hour); each temperature gradient lasted 6h. Replace the water in the cells with the ethanol solution.
[0222] 3. Centrifuge at 3000g at 4°C for 30s, remove the ethanol solution, and then wash three times with NIbuffer buffer placed on ice, after washing, follow the steps of 2×10 6 Add 20 mL of each cell to pre-cooled (pre-cooled on ice) NIbuffer to form a suspension, and shake gently f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com