SNP molecular markers linked with rice sodium and potassium ion absorption QTL and application thereof
A technology of molecular markers and ion absorption, applied in the field of molecular biology and crop genetics and breeding, can solve the problem of lack of molecular markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
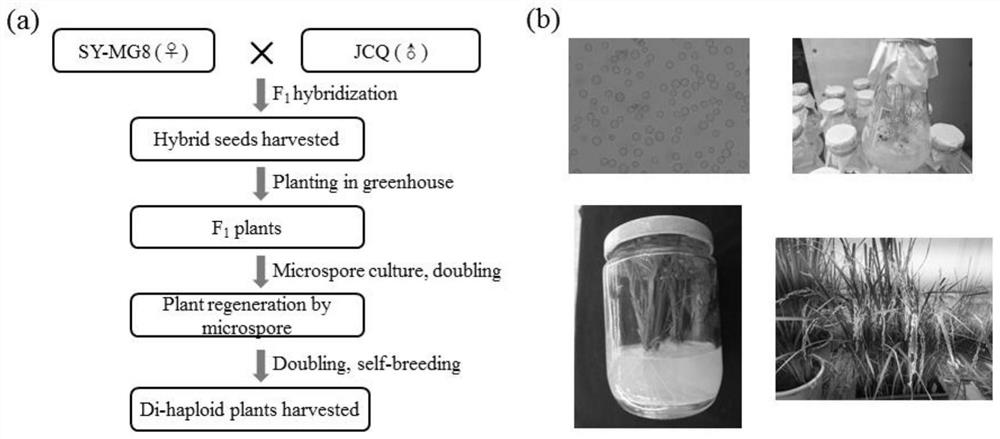
[0058] Example 1: Phenotypic identification of parental materials and rice F 1 Generation microspore DH population construction
[0059] (1) Based on the parents "Jiucaiqing" and "Shenyou-MG8", field hybridization, bagging, and harvesting hybrid F 1 generation seeds, grown in pots in the greenhouse, to obtain F 1 Generation plant, microspore culture, doubling, obtain microspore regenerated plant, doubling, selfing and fruiting, seed harvesting, obtain altogether 39 rice strains ( figure 1 ).
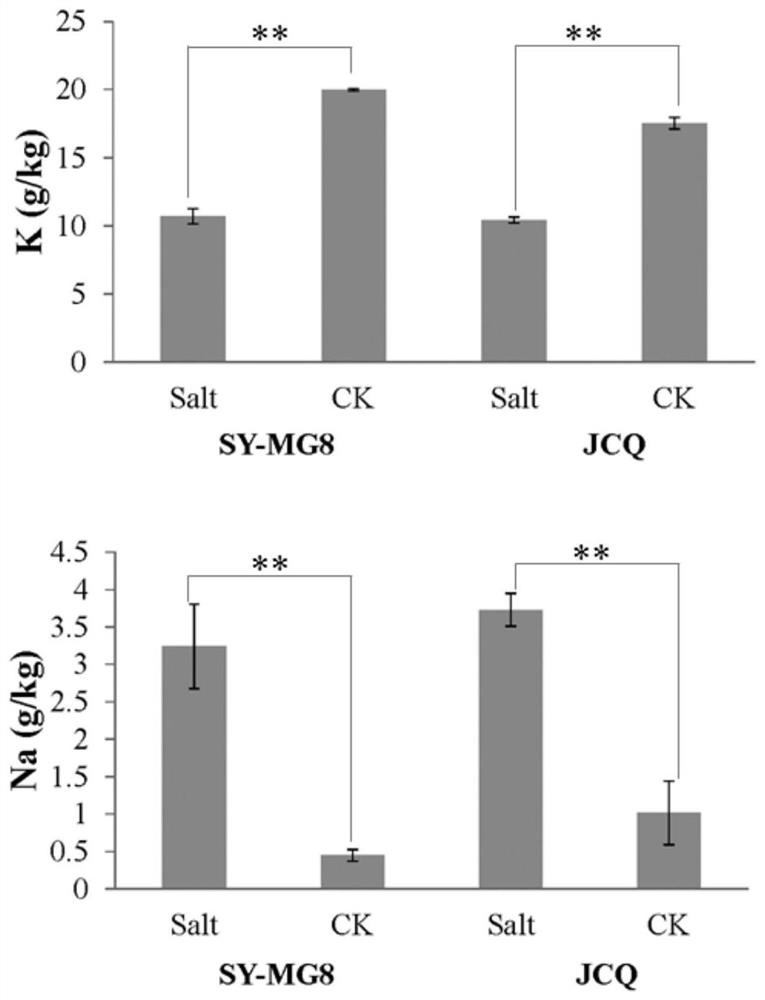
[0060](2) Using a hydroponic experiment, carry out salt stress treatment on the parents, adjust the salt concentration of the salt pond, measure the concentration of the salt pond every 2-3 days, and adjust it to 0.3%. After 30 days of salt stress, the second leaves of the plants in the salt pond were taken, and 3 plants were taken for each material. like figure 2 As shown, under salt stress, the K of variety SY-MG8 + =10.7±0.56(g / kg), Na + =3.24±0.15, K of JCQ + =10.42±0.08, Na...
Embodiment 2
[0061] Example 2: Phenotypic identification of rice DH progeny sodium and potassium ion absorption
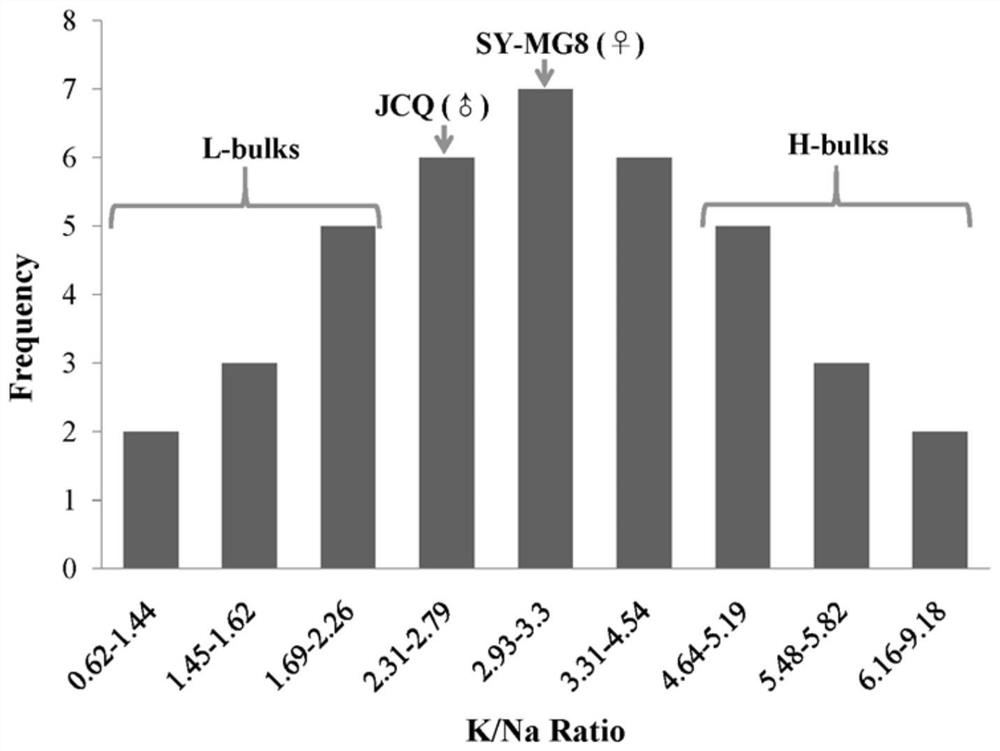
[0062] K of DH population under salt stress treatment + / Na + Ion ratio was investigated, and the K under salt treatment was screened out + / Na + The 10 lines with the highest and lowest ion ratios. like image 3 As shown, the DH population K + / Na + The ratio distribution ranged from 0.62 to 9.18. In the two extreme mixed pools derived from the DH population, divided into high K + / Na + Than groups (H-bulks) and lower K + / Na + Compared with the population (L-bulks), the difference between the two extreme pools was significant (P=2.52×10 -8 ). The two populations of H-bulks and L-bulks each included 10 DH lines, and each DH line contained 6 individuals. DH strain K in L-bulks + / Na + The value is between 0.62 and 2.26, the DH strain K in L-bulks + / Na + The value is between 4.64 and 9.18, and the remaining 19 DH strains K + / Na + The value is between 2.31 an...
Embodiment 3
[0063] Example 3: Comparative analysis of the quality of sequencing data
[0064] (1) DNA was extracted from each strain, and sampling was repeated three times for each strain. Use the NanoDrop 2000 (cThermoScientific) instrument to measure the DNA concentration, and mix in equal amounts for the construction of high K + / Na + Than mixed pool (H-bulks) and lower K + / Na + Compare pools (L-bulks) for library preparation and detection.
[0065] (2) Library construction and sequencing: Use ultrasonic waves to fragment the parental and pooled DNA sequences to form random fragments, perform end repair on the fragmented DNA in sequence, add A to the 3' end, connect the sequencing adapter, and then use magnetic beads to absorb and enrich Fragments with a genome length of about 400 bp were amplified by PCR to form a sequencing library. The library that meets the quality inspection standard is sequenced by Illumina HiSeq, the average sequencing depth of the parents is 28×, and the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com