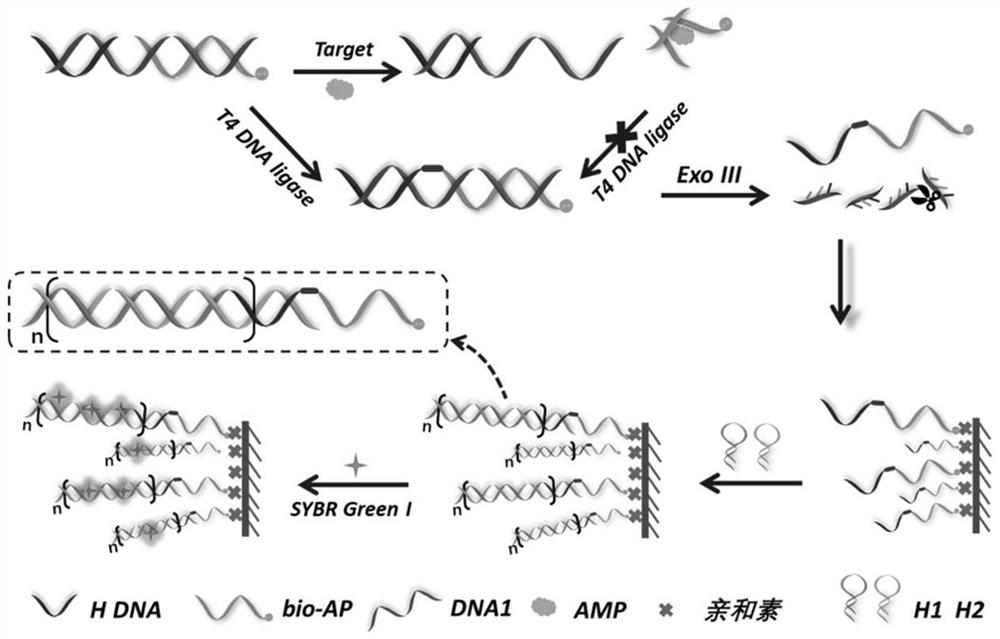
A dual-enzyme-mediated cascade signal amplification aptasensor for ampicillin detection
An aptamer sensor and ampicillin technology are applied in the determination/inspection of microorganisms, instruments, measuring devices, etc., which can solve the problems of expensive reagents, precision instruments and technicians, low sensitivity and specificity, and cumbersome analysis process, etc., to achieve High binding affinity, strong scalability, and the effect of overcoming steric hindrance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 Preparation of Probes
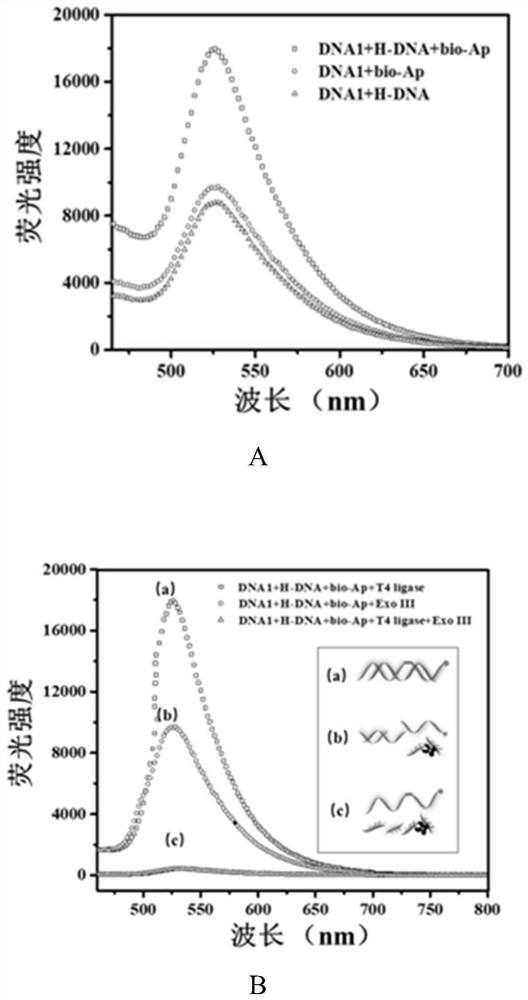
[0047] Take 50 μL of 10 μM DNA1, bio-Apt, and H-DNA priming strands, respectively, and place them in a metal thermostatic oscillator, and activate at 70 °C for 10 min. After mixing evenly, incubate at 4 °C for 2 h to complete the preparation of probes, and store at 4 °C for future use. Then, 10 μL of SYBR Green I with a final concentration of 5× was added to it, incubated at 4 °C for 30 min, and the fluorescence intensity of the sample at 520 nm under excitation light of 490 nm was detected. In order to verify the hybridization between DNA1, bio-Apt and H-DNA. The following samples were selected: a) DNA1 and bio-Apt in equal volume and concentration (10 μL, 10 μM), and 10 μL water; b) DNA1 and H-DNA in equal volume and concentration (10 μL, 10 μM), and 10 μL water. The mixture was incubated at 4 °C for 2 h. Afterwards, 10 μL of SYBR Green I with a final concentration of 5× was added to the two systems respectively, incubated at 4 °C...
Embodiment 2T4
[0049] Example 2 Activity verification of T4 ligase and Exo III
[0050] Test process: a) Take 10 μL of 100 U / mL T4 ligase and 100 μL of detection probe, add 90 μL of PBS buffer solution to mix, incubate at 37 °C for 30 min, and then inactivate at 70 °C for 20 min. After cooling, 10 μL of SYBR Green I with a final concentration of 5× was added and incubated at 4 °C for 30 min. b) Take 10 μL of 100 U / mL Exo III and 100 μL of detection probe, add 90 μL of PBS buffer solution to mix, incubate at 37 °C for 30 min, and then inactivate at 70 °C for 20 min. After cooling, 10 μL of SYBR Green I with a final concentration of 5× was added and incubated at 4 °C for 30 min. c) Take 10 μL of 100U / mL T4 ligase and 100 μL of detection probe, and incubate at 16 °C for 30 min. Add 10 μL 100 U / mL ExoIII and mix with 80 μL PBS buffer solution, incubate at 37 °C for 30 min, and then inactivate at 70 °C for 20 min. After cooling, 10 μL of SYBR Green I with a final concentration of 5× was added ...
Embodiment 3
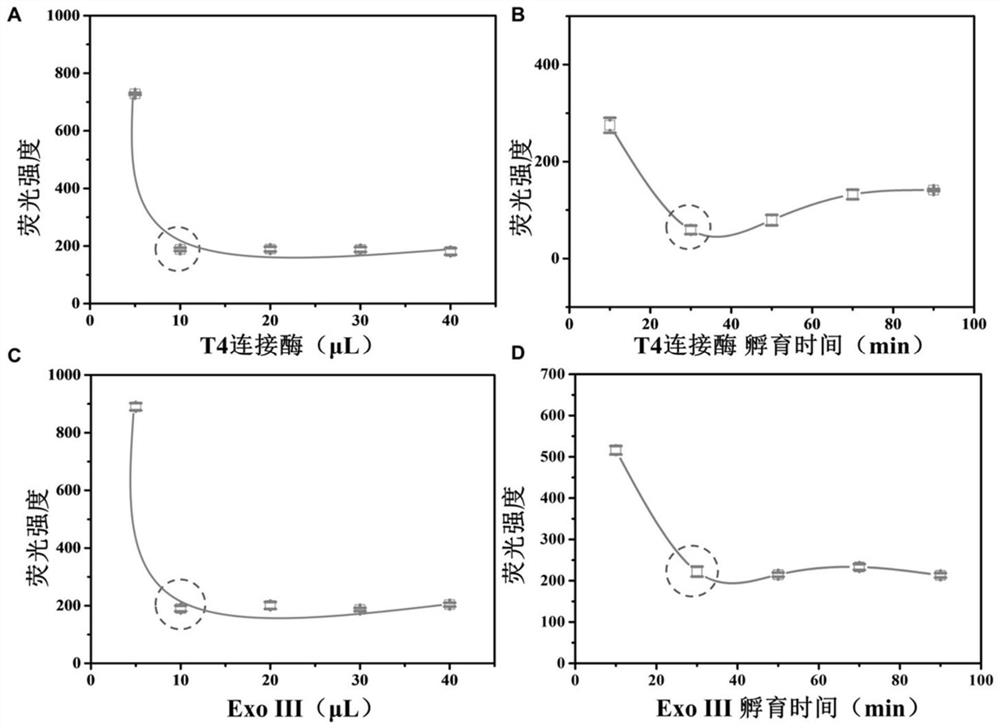
[0052] Example 3 Optimization of experimental conditions for double-enzyme-mediated DNA degradation reaction
[0053] Take 5, 10, 20, 30 and 40 μL of 100 U / mL T4 ligase and 60 μL of detection probe, respectively, and incubate at 16 °C for 30 min. Add 10 μL of 100 U / mL Exo III and mix with 80 μL of PBS buffer, incubate at 37 °C for 30 min, and then inactivate at 70 °C for 20 min. After cooling, 10 μL of SYBR Green I with a final concentration of 5× was added and incubated at 4 °C for 30 min. Detect the fluorescence intensity value of the solution at 520 nm.
[0054] see image 3 A, When the volume of T4 ligase is 10 μL, the fluorescence intensity of the solution reaches a steady state, which is the optimal reaction volume.
[0055] Take 10 μL of 100 U / mL T4 ligase and 60 μL of detection probe, respectively, and incubate at 16 °C for 10, 30, 50, 70 and 90 min. Add 10 μL of 100 U / mL Exo III and mix with 80 μL of PBS buffer, incubate at 37 °C for 30 min, and then inactivate at...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com