Method for efficiently knocking out TIGIT gene in NK cell
A kind of NK cell and high-efficiency technology, applied in the field of gene editing, has achieved obvious application prospects and clinical application value, high knockout efficiency and strong anti-tumor activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] A method for efficiently knocking out the TIGIT gene in NK cells, comprising the following specific steps:
[0037] 1. Expansion and culture of NK cells
[0038] 1) Take out the frozen human peripheral blood mononuclear cells (PBMC) from liquid nitrogen, and thaw them rapidly in a water bath at 37°C;
[0039]2) Add 4 mL of RPMI-1640 complete culture solution containing 10% FBS and 1% penicillin / streptomycin double antibody to a new 15 mL centrifuge tube, and transfer 1 mL of PBMC suspension to the 15 mL centrifuge tube;
[0040] 3) Centrifuge at 250×g for 5 minutes at room temperature;
[0041] 4) Discard the supernatant and resuspend the cells in 1 mL RPMI-1640 culture medium;
[0042] 5) Add 19 mL of RPMI-1640 culture medium into a new 75 mL cell culture flask, and transfer the above cell suspension to the culture flask;
[0043] 6) Add human recombinant IL-2 protein to the culture flask, the final concentration is 200U / mL;
[0044] 7) Place the culture flask at 3...
Embodiment 2
[0075] Detection of gene knockout efficiency by flow cytometry:
[0076] 1) Collect wild-type or NK cells after knocking out the TIGIT gene and wash twice with 1×PBS containing 1% fetal bovine serum (FBS);
[0077] 2) Resuspend the cells with PBS containing 1% FBS and count, and adjust the cell concentration to 3×10 6 a / mL;
[0078] 3) Add 50 μL of the above cell suspension into a new 1.5 mL centrifuge tube, add 1 μL of PE-anti TIGTI antibody, and incubate at 4°C in the dark for 30 min;
[0079] 4) Wash twice with 1% FBS 1×PBS;
[0080] 5) Resuspend the cells with 300 μL 1% FBS 1×PBS and perform flow cytometry detection.
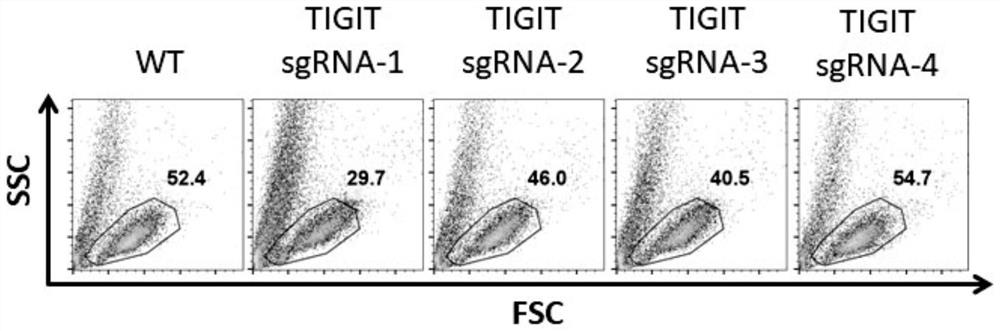
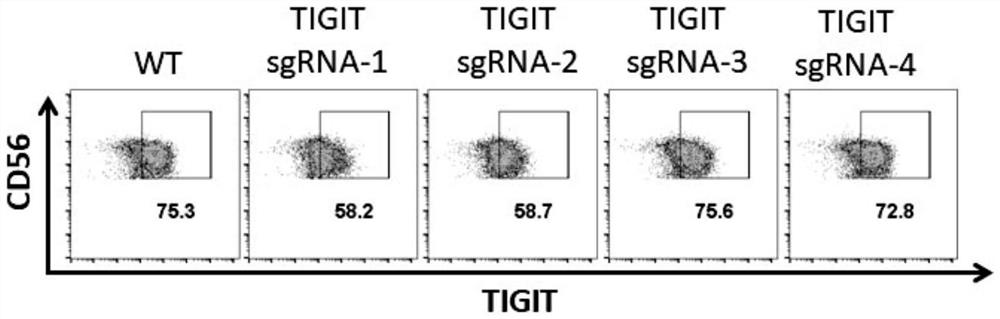
[0081] Test results such as Figure 2A and 2B shown by Figure 2A and 2B The results shown show that sgRNA-1 and 2 have higher knockout efficiency than sgRNA-3 and 4, and sgRNA-2 has less effect on cell viability than sgRNA-1, as shown in Table 1 for details:
[0082] Table 1 TIGIT knockout efficiency detected by flow cytometry
[0083] na...
Embodiment 3
[0086] The phosphorylation-modified gRNA targeting TIGIT was purchased from GenScript Biotechnology Co., Ltd. The specific phosphorylation sites are: 3 thiol and methoxy modifications at the 3' end and 5' end respectively.
[0087] The specific sequence is TIGIT-2: CTGGTGTCTCCTCCTGATCT
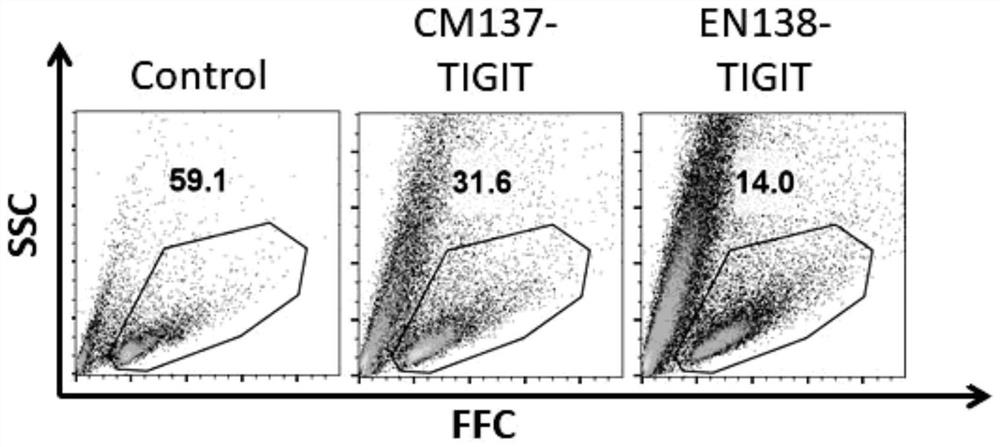
[0088] Figure 3A It shows the comparison of the cell activity of NK cells after the unmodified and phosphorylated sgRNA-2 sequences targeting TIGIT were electroporated by the CM137 program, and the WT group was NK cells electroporated with only Cas9 protein but not electroporated sgRNA ;
[0089] Figure 3B It shows the comparison of the knockout efficiency of TIGIT gene after electroporation of unmodified and phosphorylated sgRNA-2 sequences targeting TIGIT, where CD56 is a surface marker of human NK cells, and the WT group is only electroporated with Cas9 NK cells with protein but not electroporated sgRNA;
[0090] Figure 3C Comparison of sequencing analysis results of unmodified and ph...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com