Hybridoma cell strain 3G7 1B10, anti-GII.4 type norovirus P protein monoclonal antibody and application
A technology of 3G71B10 and hybridoma cell lines, applied in the direction of antiviral immunoglobulin, immunoglobulin, chemical instruments and methods, etc., can solve the problems of unsatisfactory human norovirus, missed detection, lack of cross-reactivity, etc. Achieve good pairing effect, good cross-reactivity and clear results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] The preparation of embodiment 1 hybridoma cell line and the acquisition of monoclonal antibody
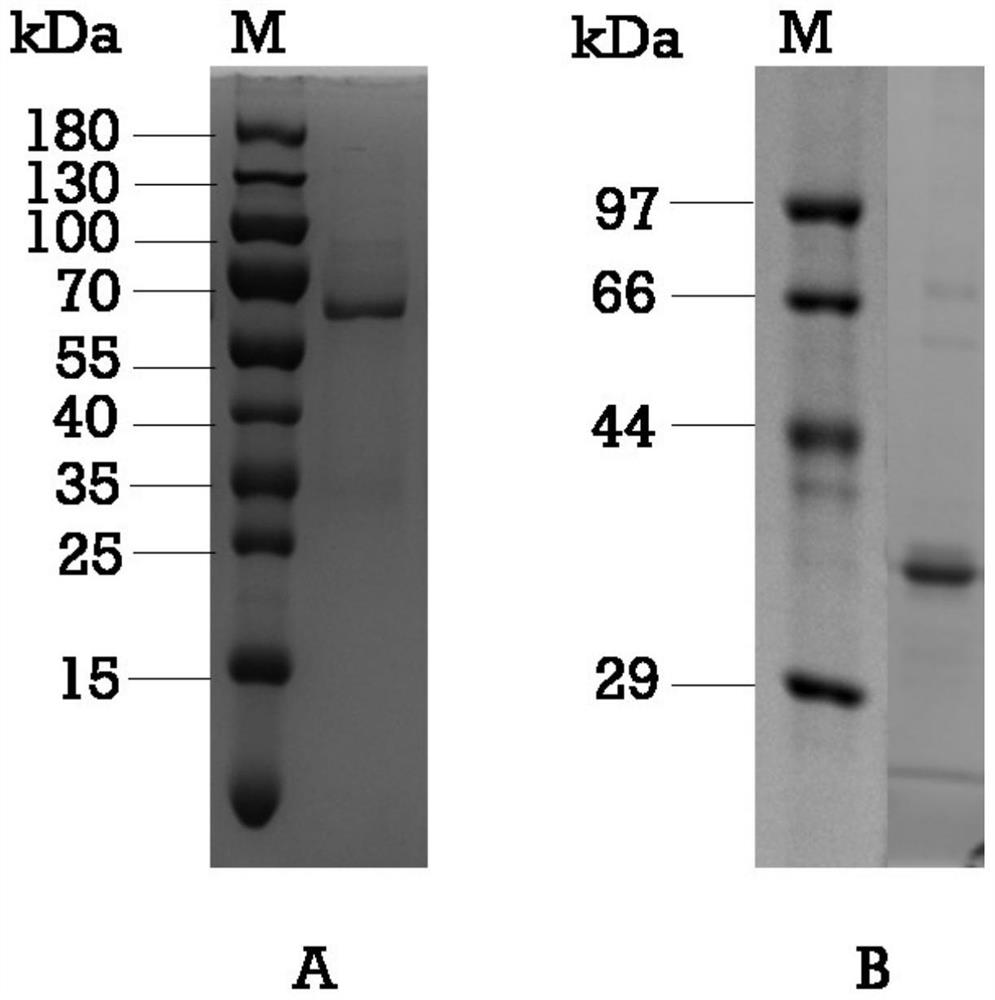
[0025] 1. Preparation of GII.4 type norovirus P protein immunogen
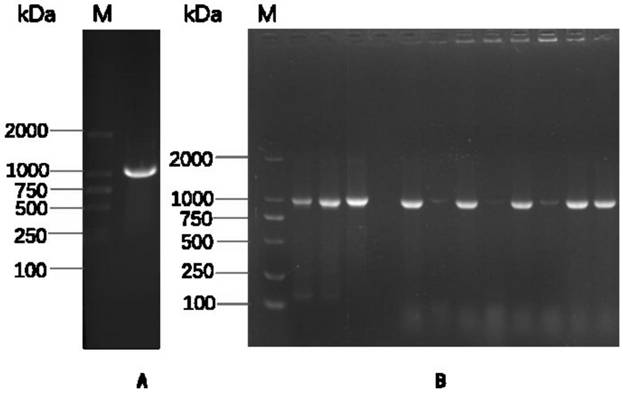
[0026] Amplification primers were designed according to the GII.4-2012 type GZ2014-L307 strain (GenBank KT202798) P region, the upstream primer P1 and the downstream primer P2 respectively introduced BamHI and EcoRI restriction sites (underlined part), and the downstream primer 5' Adding the CDCRGDCFC amino acid sequence (italics) at the end can promote the formation of Norovirus P particles, as shown in Table 1. The GII.4-2012 type GZ2014-L307 strain cDNA was used as a template for PCR amplification, and the amplified product and the pGEX-4T-1 vector (GE General Healthcare, USA) were subjected to BamHI and EcoRI double enzyme digestion, respectively. The amplified product was connected to the pGEX-4T-1 vector, transformed into Escherichia coli BL21 competent cells, and verified by colony PCR and sequencing...
Embodiment 2
[0041] The establishment of embodiment 2 double antibody sandwich ELISA method
[0042] 1. Screening of paired antibodies and determination of optimal working concentration
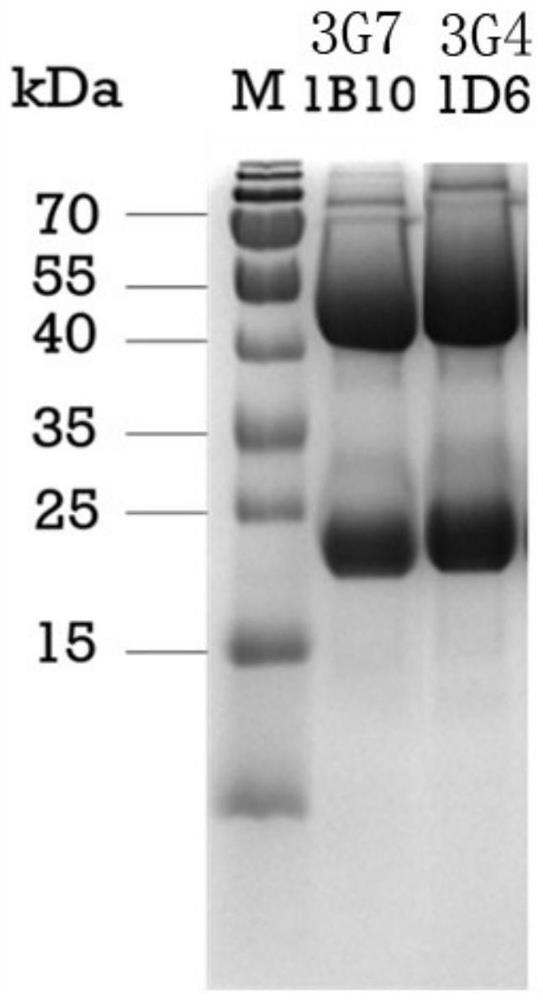
[0043] First, the two monoclonal antibodies were labeled according to the instructions of the horseradish peroxidase (HRP) coupling labeling kit, and the sensitivity of the labeled antibody was measured by direct ELISA, and the detection antibody was determined. Using the method of double-antibody sandwich ELISA, 1 μg / mL GII.4 P particles were used as the detection antigen, and the identified HRP-labeled monoclonal antibodies (HRP-3G7 1B10, HRP-3G4 1D6) were used as the detection antibodies, and the other two monoclonal antibodies ( 3G7 1B10, 3G4 1D6) were paired with the capture antibody respectively, and the best paired antibody and the optimal working concentration were determined. The results showed that HRP-labeled 3G7 1B10 antibody (HRP-3G7 1B10) had the best sensitivity, so HRP-3G7 1B10 was select...
Embodiment 3
[0046] The performance test of embodiment 3 method
[0047] The performance test of the double-antibody sandwich ELISA method established in Example 2 is as follows:
[0048] 1. Specificity test
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com