Substrate combination, detection reagent and detection kit for detecting escherichia coli and/or shigella
A technology for Escherichia coli and Shigella, which is applied to measurement devices, material analysis, instruments and other directions by observing the impact on chemical indicators, can solve the problems of inability to identify Shigella, time-consuming, and complicated steps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] A clinical strain, the result of mass spectrometry identification is Escherichia coli, the operation steps are as follows:
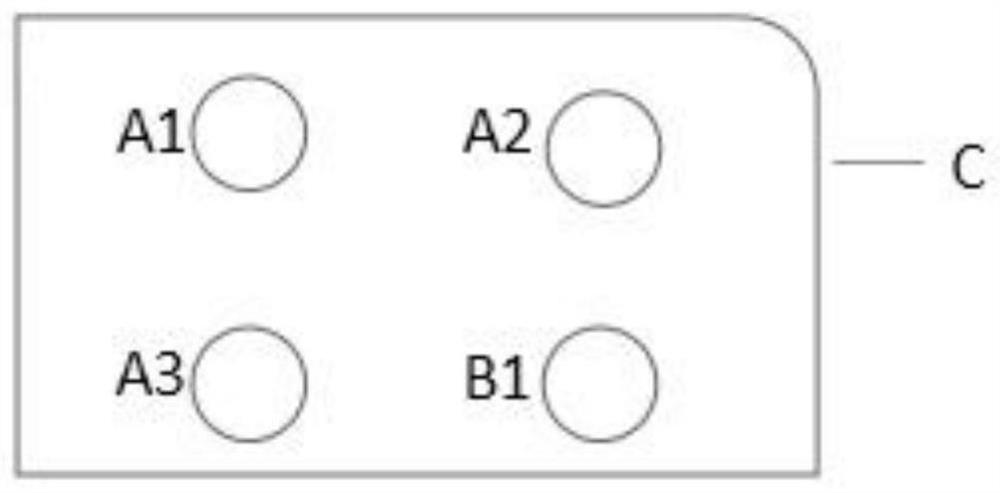
[0071] 1. Board preparation, as follows:
[0072] Well 1: 0.8% mannitol, 1.4% phenol red, phosphate buffer pH7.2
[0073] Well 2: 0.8% ONPG (2-nitrophenyl-β-D-galactopyranoside), phosphate buffer pH7.0
[0074] Well 3: 0.8% sorbitol, 1.4% phenol red, phosphate buffer pH7.2
[0075] Well 4: 0.5% glycerol, 1.4% phenol red, phosphate buffer pH7.2
[0076] 12ul of each coating solution was coated on a four-well plate card and used after natural drying
[0077] 2. Preparation of bacterial suspension: use a turbidimeter to prepare 2.5 liters of Maibella bacteria from the strain to be tested, add 100ul per well to the above-mentioned plate, and place the plate in a 37°C incubator for 1.5h after adding the sample.
[0078] 3. Interpretation of results: Take out the board, record the color of the reaction well and the corresponding yin and yang results...
Embodiment 2
[0086] A clinical strain, the result of mass spectrometry identification is Escherichia coli, the operation steps are as follows:
[0087] 1. Board preparation, as follows:
[0088] Well 1: 1.2% mannitol, 1.7% phenol red, Tris buffer pH7.4
[0089] Well 2: 1.2% ONPG (2-nitrophenyl-β-D-galactopyranoside), Tris buffer pH8.0
[0090] Well 3: 1.2% sorbitol, 1.7% phenol red, Tris buffer pH7.4
[0091] Well 4: 1% glycerol, 1.7% phenol red, Tris buffer pH7.4
[0092] 8ul of each coating solution was coated on a four-well plate, and used after natural drying
[0093] 2. Preparation of bacterial suspension: use a turbidimeter to prepare 3.0 strains of Maibella bacteria from the strain to be tested, add 100ul per well to the above-mentioned plate, and place the plate in a 37°C incubator for 1 hour after adding the sample.
[0094] 3. Interpretation of results: Take out the board, record the color of the reaction well and the corresponding yin and yang results
[0095] Well 1: yello...
Embodiment 3
[0102] A clinical strain, the result of mass spectrometry identification is Escherichia coli, the operation steps are as follows:
[0103] 1. Board preparation, as follows:
[0104] Well 1: 1% mannitol, 1.6% phenol red, phosphate buffer pH7.3
[0105] Well 2: 1% ONPG (2-nitrophenyl-β-D-galactopyranoside), phosphate buffer pH7.5
[0106] Well 3: 1% sorbitol, 1.6% phenol red, phosphate buffer pH7.3
[0107] Well 4: 0.8% glycerol, 1.6% phenol red, phosphate buffer pH7.3
[0108] 10ul of each coating solution was coated on a four-well plate card, and used after natural drying
[0109] 2. Preparation of bacterial suspension: use a turbidimeter to prepare 2.8 molasses strains from the strain to be tested, add 100ul per well to the above-mentioned plate, and place the plate in a 37°C incubator for 2 hours after adding the sample.
[0110] 3. Interpretation of results: Take out the board, record the color of the reaction well and the corresponding yin and yang results
[0111] Ho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com