Mitochondrial microsatellite marker primers for pigeon genetic diversity analysis and detection method
A technology of genetic diversity and microsatellite markers, applied in the field of mitochondrial microsatellite marker primers and detection, can solve the problems of no microsatellite molecular markers and applications, and achieve accurate kinship analysis data, large numbers, and good polymorphism Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026]Example 1 Research on genetic diversity of white king pigeon
[0027](1) Extract genomic DNA from 36 white king pigeon feathers;
[0028](2) Using the DNA extracted in step (1) as a template, synthesize forward and reverse primers, and label the 5'end of the forward primer with FAM fluorescent dye to perform PCR amplification reaction;
[0029]The sequence of the forward and reverse primers is as follows:
[0030]Forward primer: 5'-TTTCCTATGATTACCACTGG-3'; SEQ ID NO.1;
[0031]Reverse primer: 5'-TGTAAGCTACGGGGACG-3'; SEQ ID NO. 2;
[0032]The PCR reaction system is: 2×PCR Mix (Nanjing Novez Biotech Co., Ltd.) 25μL, 10μmol / L forward and reverse primers each 1μL, 50-100μg / ml template DNA 2μL, ultrapure water 21μL;
[0033]The PCR reaction program is: 95°C 5min; 95°C 30s, 59°C 30s, 72°C 60s, 35 cycles; 72°C 10min;
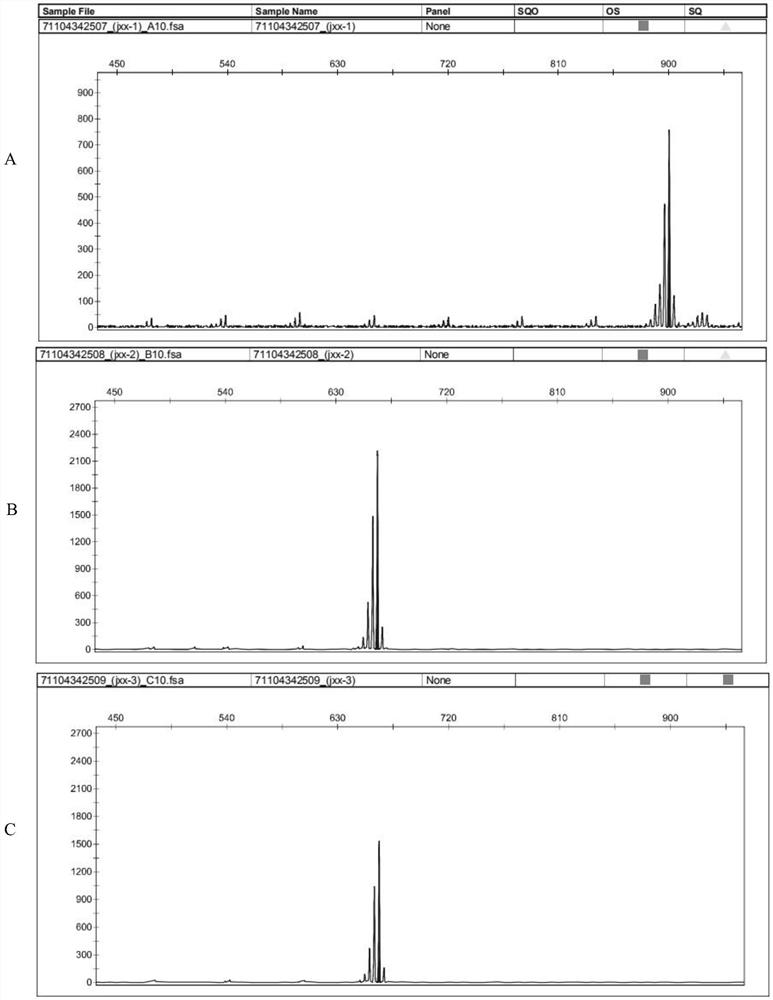
[0034](3) The PCR product is genotyped on the ABI 3730 analyzer;
[0035](4) Use Cervus3.0 software to analyze the number of alleles (NA), Observedheterozygosity (Observedheterozygosity, HO), E...
Embodiment 2
[0039]Example 2 Verification of Maternal Relationship of Silver King Pigeon
[0040](1) Extract the genomic DNA from the feathers of a litter of silver king pigeons (male pigeons, female pigeons, 2 squabs);
[0041](2) Using the DNA extracted in step (1) as a template, synthesize forward and reverse primers, and label the 5'end of the forward primer with FAM fluorescent dye to perform PCR amplification reaction;
[0042]The sequence of the forward and reverse primers is as follows:
[0043]Forward primer: 5'-TTTCCTATGATTACCACTGG-3'; SEQ ID NO.1;
[0044]Reverse primer: 5'-TGTAAGCTACGGGGACG-3'; SEQ ID NO. 2;
[0045]The PCR reaction system is: 2×PCR Mix (Nanjing Novez Biotech Co., Ltd.) 25μL, 10μmol / L forward and reverse primers each 1μL, 50-100μg / ml template DNA 2μL, ultrapure water 21μL;
[0046]The PCR reaction program is: 95°C 5min; 95°C 30s, 59°C 30s, 72°C 60s, 35 cycles; 72°C 10min;
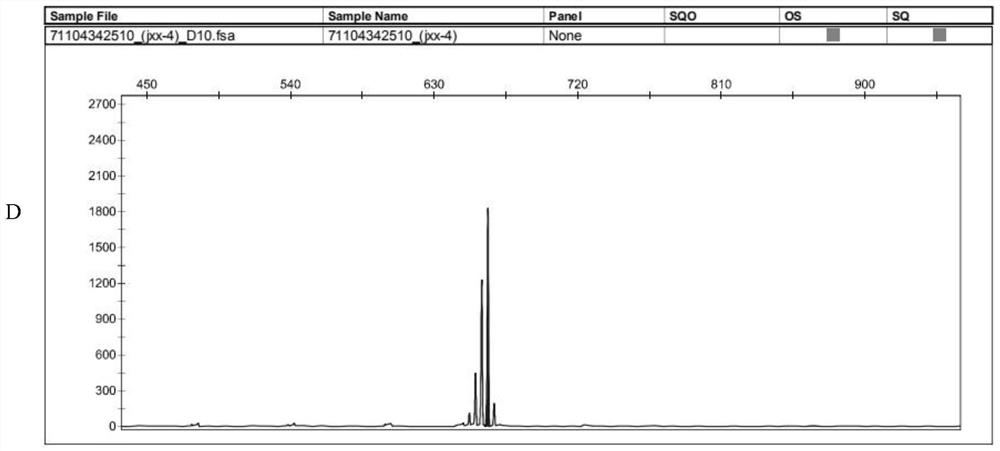
[0047](3) The PCR product was genotyped on the ABI 3730 analyzer; the results showed that the bands of the two squabs ...
Embodiment 3
[0048]Example 3 Verification of Maternal Relationship of Taishen Pigeon
[0049](1) Extract the genomic DNA from the blood of a litter of Taishen pigeons (male pigeons, female pigeons, 2 young pigeons);
[0050](2) Using the DNA extracted in step (1) as a template, synthesize forward and reverse primers, and perform PCR amplification reaction;
[0051]The sequence of the forward and reverse primers is as follows:
[0052]Forward primer: 5'-TTTCCTATGATTACCACTGG-3'; SEQ ID NO.1;
[0053]Reverse primer: 5'-TGTAAGCTACGGGGACG-3'; SEQ ID NO. 2;
[0054]The PCR reaction system is: 2×PCR Mix (Nanjing Novez Biotech Co., Ltd.) 25μL, 10μmol / L forward and reverse primers each 1μL, 50-100μg / ml template DNA 2μL, ultrapure water 21μL;
[0055]The PCR reaction program is: 95°C 5min; 95°C 30s, 59°C 30s, 72°C 60s, 35 cycles; 72°C 10min;
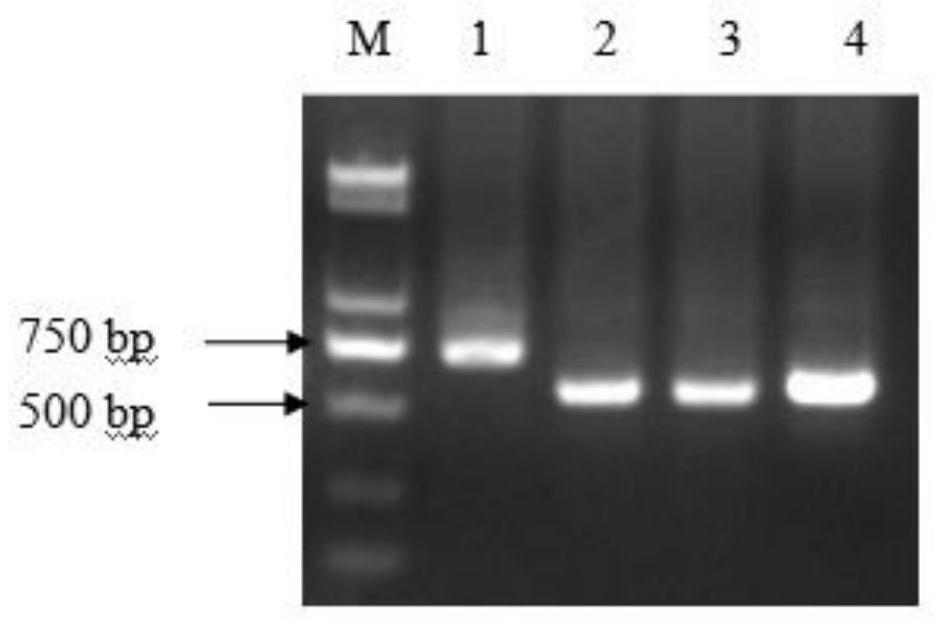
[0056](3) The PCR products were subjected to agarose gel electrophoresis; the results showed that the bands of the two pigeons were consistent with the hens, but not consistent with the co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com