Method for synthesizing S-adenosylmethionine from DL-methionine by modifying saccharomyces cerevisiae based on CRISPR technology
A technology of adenosylmethionine and Saccharomyces cerevisiae, applied in the direction of microorganism-based methods, other methods of inserting foreign genetic materials, recombinant DNA technology, etc., can solve the problems of less development and research, and achieve the effect of reducing costs and improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
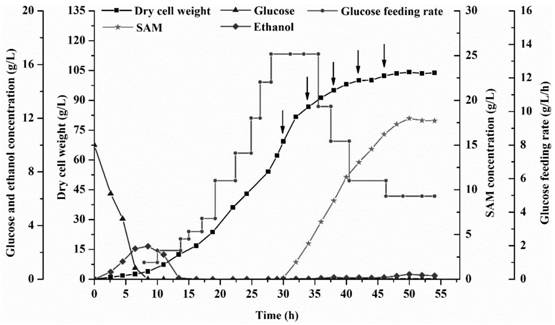
Image
Examples
Embodiment 1
[0028] The selection of CRISPR / Cas9 system in the industrial Saccharomyces cerevisiae of embodiment 1
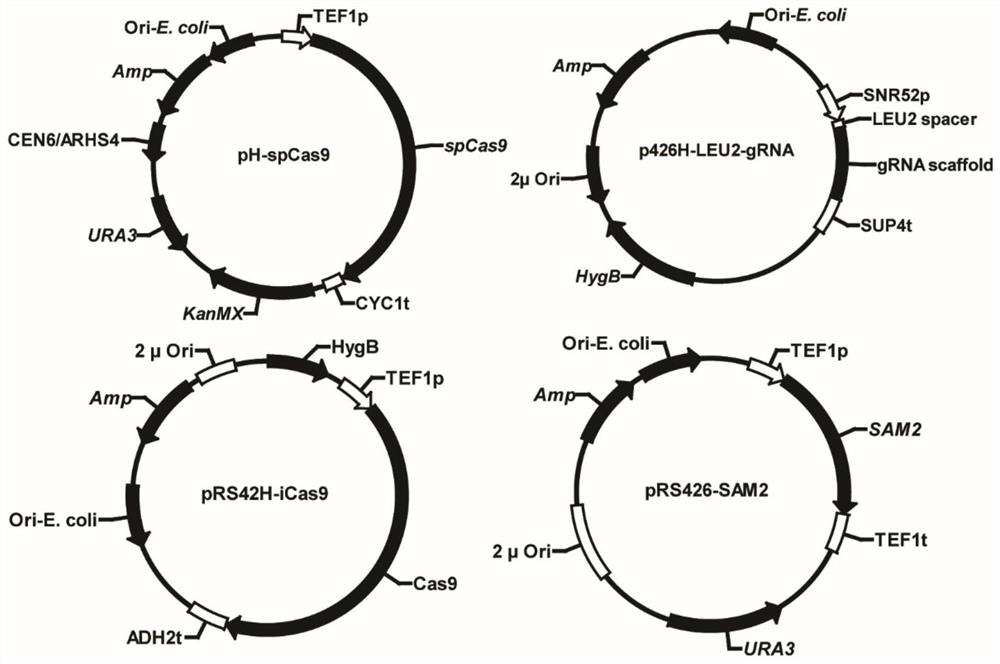
[0029] In this example, in order to establish a suitable genome editing CRISPR system in diploid industrial Saccharomyces cerevisiae HD, the most reported CRISPR / Cas9 dual-plasmid system in S. CEN / ARS element) expresses Cas9 (pH-spCas9) (SEQ ID No.1), and the multi-copy plasmid (2μ plasmid) expresses gRNA (p426H-LEU2-gRNA) (SEQ ID No.2). Leucine deficiency occurs due to inactivation of the LEU2 gene on the S. cerevisiae genome, and only simultaneous inactivation of all alleles in polyploid yeast results in a leucine-deficient phenotype.
[0030] Using the plasmid pRS426-SAM2 (SEQ ID No.3) as a template, primers Leu-SAM2-For (SEQ ID No.4) and Leu-SAM2-Rev (SEQ ID No.5) were used to amplify 40 bp of the LEU2 gene at both ends. The fragment of the SAM2 gene expression cassette of the source sequence was used as a homology repair template to knock in the LEU2 gene to examine th...
Embodiment 2
[0034] Example 2 Optimization of gRNA plasmid and repair template DNA concentration
[0035] During the transformation process, different concentrations of gRNA and repair templates have a great influence on the efficiency of gene editing. Therefore, in this example, different concentrations of gRNA and repair templates were used in the process of pRS42H-iCas9 and pKan100-LEU2-gRNA-mediated knock-in of the LEU2 gene into the SAM2 expression frame. The results are shown in Table 2. When the concentration of the pKan100-LEU2-gRNA plasmid reaches 2.5 μg and the concentration of the repair template containing the SAM2 expression cassette reaches 5 μg, the gene knock-in efficiency can reach nearly 100%. Therefore, both gRNA and repair templates are added at this concentration in subsequent gene editing.
[0036] Table 2 Comparison of the efficiency of knocking LEU2 gene into SAM2 expression cassette with different gRNA plasmids and repair template concentrations
[0037]
Embodiment 3
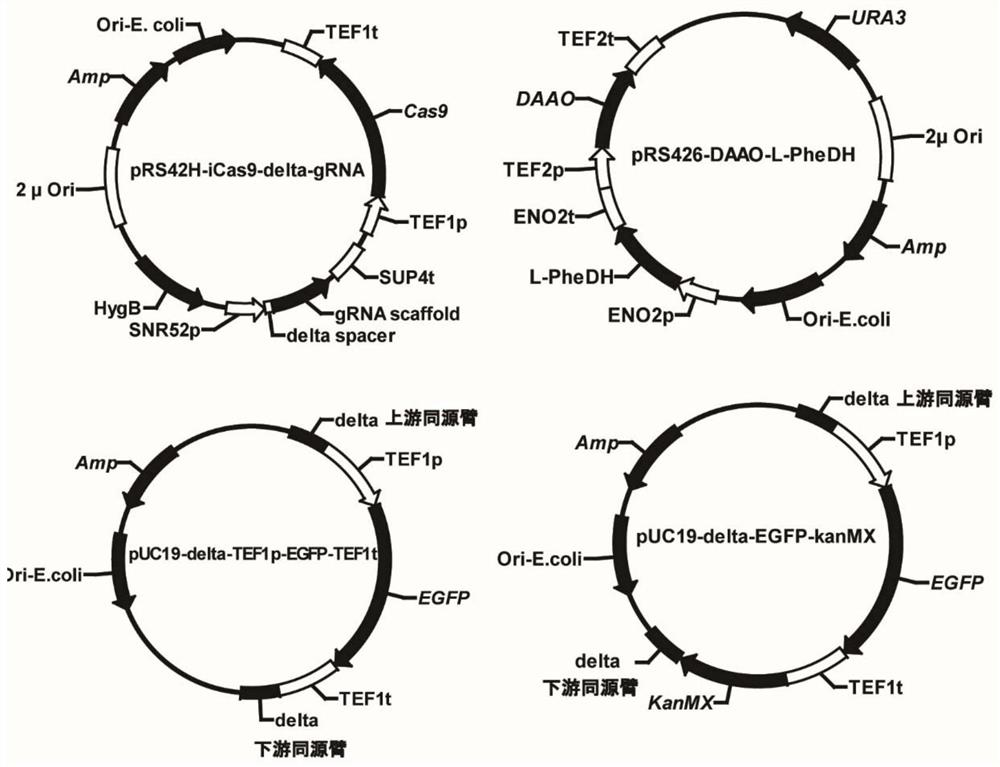
[0038] Example 3 Gene knockout and chromosome fragment deletion based on CRISPR / Cas9 system
[0039] In this example, using optimized conditions, the efficiency of gene knockout and chromosome deletion mediated by pRS42H-iCas9 and pKan100-GAL10-gRNA systems was investigated. Targeting the GAL1, GAL7 and GAL10 regions on the chromosome of Saccharomyces cerevisiae, using primers 1000bp-deletion-L-For (SEQ ID No.8) and 1000bp-deletion-L-Rev (SEQ IDNo.9), using the Saccharomyces cerevisiae genome as Template, amplifying the upstream homology arm fragment of knockout genome 1000bp, using primers 1000bp-deletion-R-For (SEQ ID No.10) and 1000bp-deletion-R-Rev (SEQ ID No.11), to Saccharomyces cerevisiae The genome is used as a template, and the downstream homology arm fragment of 1000bp of knockout genome is amplified, and the repair template DNA of 1000bp of genome knockout is obtained by fusion PCR; using the same method, using primer 2000bp-deletion-L-For (SEQ IDNo.12) , 2000bp-de...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com