Microbial marker related to colorectal cancer and application thereof
A colorectal cancer and microorganism technology, applied in the field of microorganisms, can solve the problems of intolerance or refusal of screening for colorectal cancer risk groups, the high price of detection technology, and the suffering of the subjects, and achieves good application prospects and practical significance. Accuracy and safety, high sensitivity effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Embodiment 1, the extraction of DNA sample.
[0044] (1) Collect individual feces samples, freeze them immediately, and put them on ice before the experiment;
[0045] (2) Weigh 200 mg of fixed feces into a 2 mL centrifuge tube, add 800 μL of fecal DNA extraction buffer, shake and mix well for 5 min, and centrifuge at 1800 g for 1 min;
[0046] (3) Take 50 μL of the suspension into a 1.5 mL centrifuge tube, add 800 μL of lysate, vortex and mix, lyse at 70°C for 5 min, centrifuge for 5 min, and transfer the supernatant to a clean 1.5 mL centrifuge tube;
[0047] (4) Add 20 μL of well-mixed magnetic beads, vortex for 20 seconds, let stand at room temperature for 4 minutes, place on a magnetic rack, let stand for 20 seconds, and absorb the supernatant;
[0048] (5) Add 500 μL of washing solution Ⅰ, vortex for 20 seconds, mix the magnetic beads, place them on a magnetic rack, let them stand for 20 seconds, and discard the supernatant;
[0049] (6) Add 750 μL of washing so...
Embodiment 2
[0053] Example 2, Quantitative Detection of Microbial Markers
[0054] The quantitative detection of microbial markers adopts the Taqman qPCR method, and the probes and primers used are shown in SEQ ID NO.01-09.
[0055] The specific steps of this embodiment will be described below by taking the TaqMan Master Mix kit product of Suzhou Xinhai Biotechnology Co., Ltd. as an example.
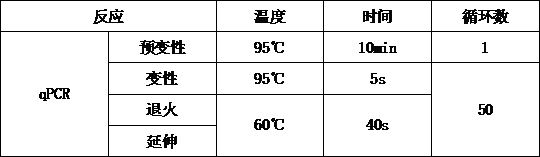
[0056] (1) Perform the reaction according to the qPCR reaction system shown in Table 1 to prepare the PCR reaction solution.
[0057] Table 1. qPCR reaction system
[0058] Reagent Dosage concentration Taqman Master Mix (2×) 12.5μL 1× PCR forward primer (10 μM) 1μL 400nM PCR reverse primer (10 μM) 1μL 400nM Probe (0.2uM) 1μL 400nM Sample DNA to be tested 1μL 1-100ng Sterilized water 8.5μL /
[0059] (2) After the PCR reaction solution is prepared, mix it upside down and centrifuge, dispense it into a 96-well PCR reaction plate, ...
Embodiment 3
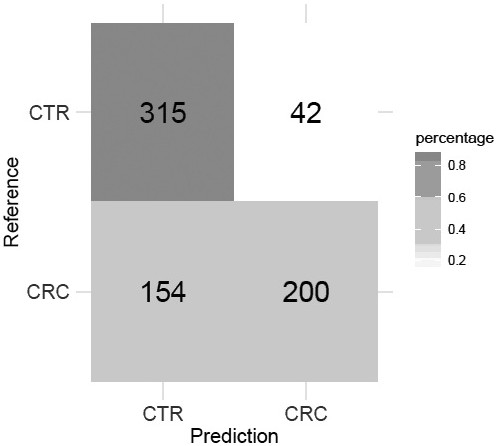
[0064] Example 3, colorectal cancer risk assessment model.
[0065] The colorectal cancer risk assessment model uses the random forest algorithm to train and test the model using the abundance of three microbial markers from 357 healthy samples and 354 colorectal cancer patient samples, and finally establishes the colorectal cancer risk assessment model, with specific steps as follows:
[0066] Step 1) Using the method described in Example 1 to extract DNA fragments from 357 healthy samples and 354 colorectal cancer samples;
[0067] Step 2) Use as described in Implementation 2 to detect the content of the target gene fragments of the three microbial markers in all samples and the gene content of the internal reference 16S rDNA;
[0068] Step 3) According to the content of the target gene fragment of the microbial marker described in step 2) and the gene content of the internal reference 16S rDNA, calculate the abundance of the corresponding microbial marker;
[0069] Step 4...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com