Attenuated pseudorabies virus QD strain by three-gene deletion
A porcine pseudorabies virus and gene deletion technology, which is applied in the direction of viruses, antiviral agents, and virus antigen components, can solve the problems of high threshold for artificial genome construction, unsuitable for rapid operation, etc., and achieve good commercial development prospects and effective The effect of immune protection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
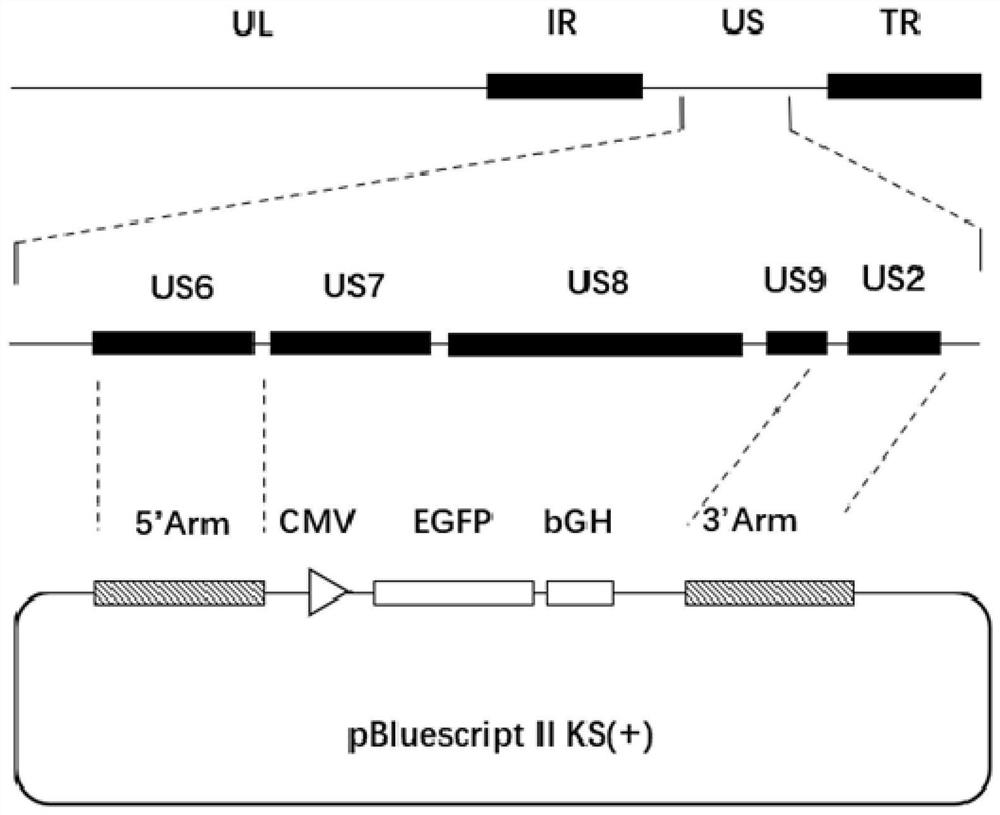
[0024] Example 1: Construction of three gene-deleted strains of gE, gI and 11K of PRV-QD
[0025] 1. Construction of recombinant vector
[0026] Taking the PRV HLJ8 strain (Genebank: KT824771) as a reference, the complete US7 (gI) gene, the complete US8 (gE) gene and the US9 (11K) gene were designed to delete the Unique Short (US) gene. Primers were designed to amplify the homology arms on both sides of the deleted gene, and the primers are shown in Table 1.
[0027] Table 1: Sequence Information Table of Primers
[0028]
[0029]
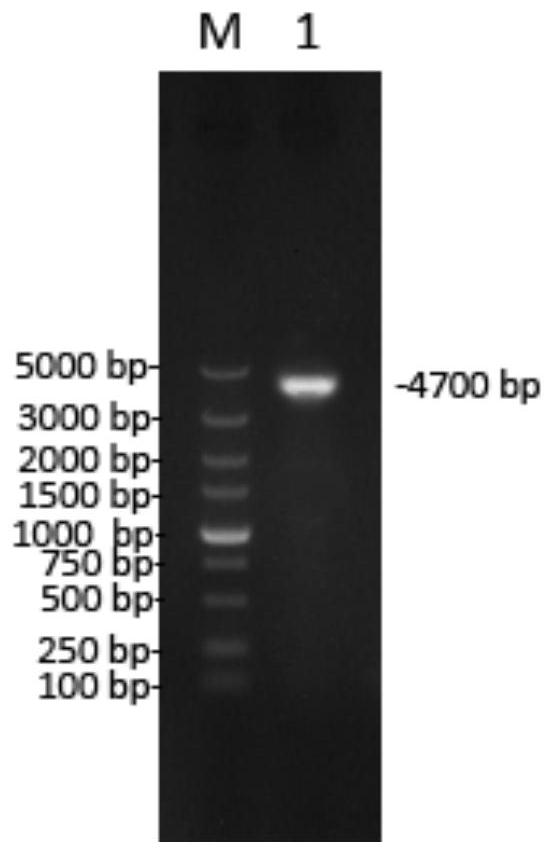
[0030] Use 5'arm F / 5'arm R and 3'arm F / 3'arm R primers to establish 50μL amplification system: 2×PrimeSTAR HS GC Buffer 25μL; dNTP (2.5mM each) 4μL; Forward primer 1μL; Reserveprimer 1μL ; PRV genomic DNA 1 μL; PrimeSTAR HS 0.5 μL; deionized water 17.5 μL. Reaction conditions: pre-denaturation at 98°C for 3 minutes; 10s at 98°C, 5s at 55°C, 1kb / min at 72°C, 30 cycles of amplification. The amplified products are the 5' arm fragment and the...
Embodiment 2
[0043] Example 2: Immunological activity of PRV-QD gE- / gI- / 11K-strain
[0044] 1. Determination of stability and growth characteristics of PRV-QD gE- / gI- / 11K-strains
[0045] PRV QD strain and PRV QD-gE- / gI- strain were respectively inoculated with monolayer PK-15 cells at the ratio of MOI 0.1, and placed in an incubator to continue culturing. The virus liquid was collected every 12 hours to measure TCID50, and the growth curve was drawn. The PRV QD-gE- / gI-strain was continuously passaged to 15 passages, and the recombined region was amplified using RetestF2 / RetestR primers and sequenced for identification.
[0046] 2. Half infection dose of PRV-QD gE- / gI- / 11K- mice
[0047] Sixty-five 6-week-old Balb / c female mice were divided into 13 groups randomly. PRV QD strain and PRV QD-gE- / gI- cell supernatant were diluted to 10-6 times with 10 times of normal saline. Mice were subcutaneously injected with 0.2 mL of virus solution of each dilution, and the negative control group wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com