Preparation method of human specific gene fluorescent probe and probe prepared by method
A fluorescent probe, specific technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of cumbersome preparation process of fluorescent probes, low labeling efficiency, and unstable labeling. , to achieve the effect of stable labeling method, simplified preparation process and high efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1: Preparation of human CDKN2A gene fluorescent probe
[0042] 1) Find the specific gene sequence
[0043] Find the CDKN2A gene on the UCSC website (http: / / genome.ucsc.edu / index.html), extend the length of the upstream and downstream of the gene location (chr9:21,967,753-994,624) by 100kb, and shield the repetitive sequence. Part of the non-repeated sequence of the post-CDKN2A gene such as figure 1 As shown, where "N" means a masked repeat sequence, and non-N means a non-repeat sequence (the complete sequence is shown in other supporting documents);
[0044] 2) Design specific primers
[0045] Referring to the design of DNA primers, search for the specific primer sequence on the obtained non-repetitive sequence, which requires that the specific primer sequence length is 20-30bp, the Tm value is 60-62°C, the distance between the two specific primers is at least 5bp, and the specific primer sequence The quantity is 1744 pieces. Import the designed specific pri...
Embodiment 2
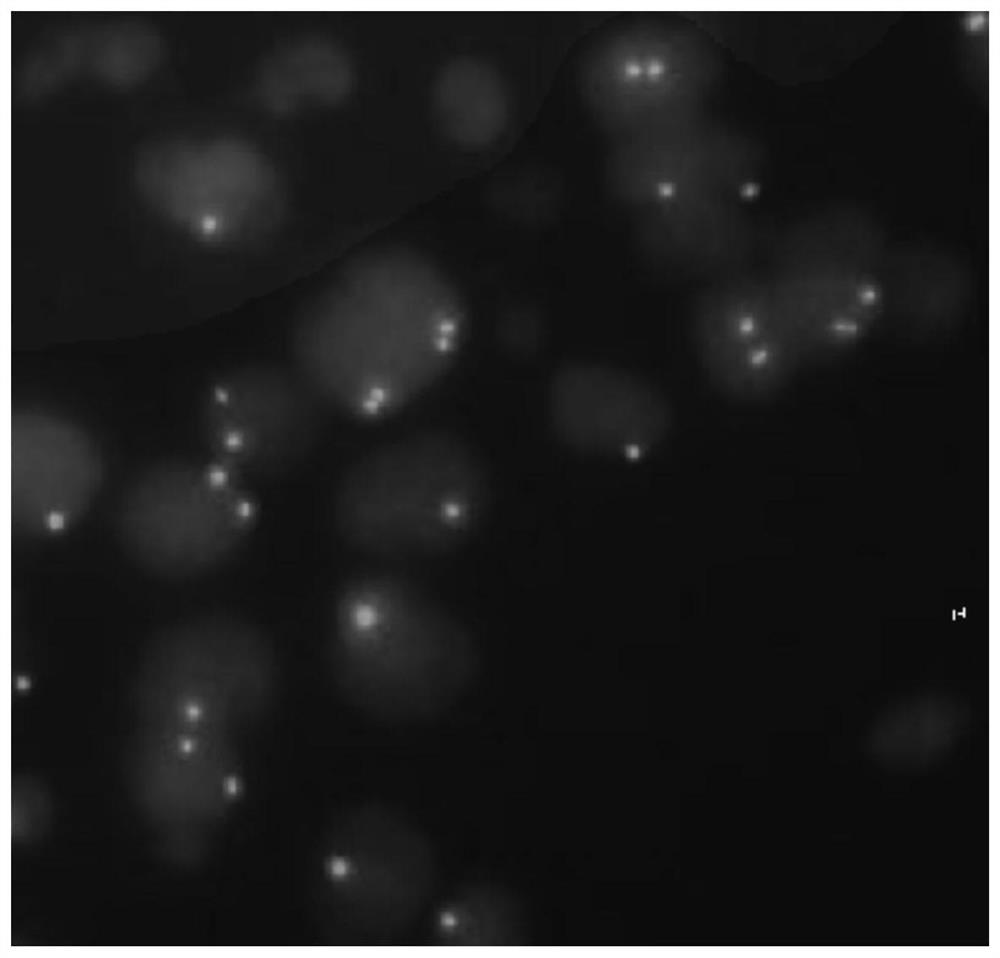
[0065] Embodiment 2: Detection of human CDKN2A gene by fluorescence in situ hybridization (FISH)
[0066] 1) Sample processing:
[0067] The peripheral blood cell droplet samples were soaked in 2×SSC solution at room temperature for 2 minutes, then soaked in 70%, 90%, and 100% ethanol at room temperature for 2 minutes to dehydrate, took out the slides, and dried them at room temperature for later use.
[0068] 2) Denatured hybridization of the sample with the probe:
[0069] Take 10ul of the probe working solution prepared in Example 1 and add it dropwise to the hybridization area, cover the cover glass while avoiding air bubbles, seal the slide along the edge of the cover glass with rubber glue, and place the slide on a hybridization instrument at 90°C Denaturation for 2 minutes, hybridization at 55°C for 30 minutes-3 hours.
[0070] 3) Washing after hybridization:
[0071] Remove the slide, remove the rubber and remove the coverslip. Place the slides in 73°C preheated wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com