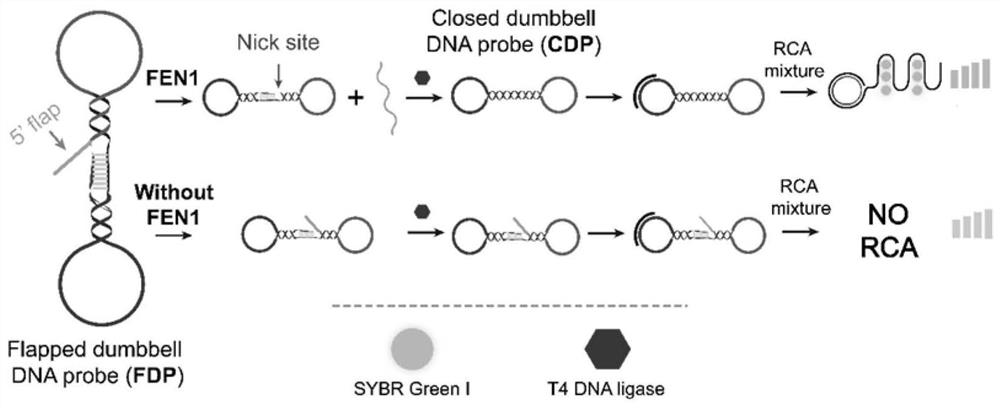
Method for detecting structure-specific nuclease FEN1 by using biosensor combining DNA ligation reaction and rolling circle amplification
A biosensor and rolling circle amplification technology, which is applied in the field of biosensor detection of structure-specific nuclease FEN1, can solve the problems that conventional detection methods cannot meet clinical needs and the development of analysis technology is weak, so as to avoid instability and improve sensitivity , the effect of effective research tools
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
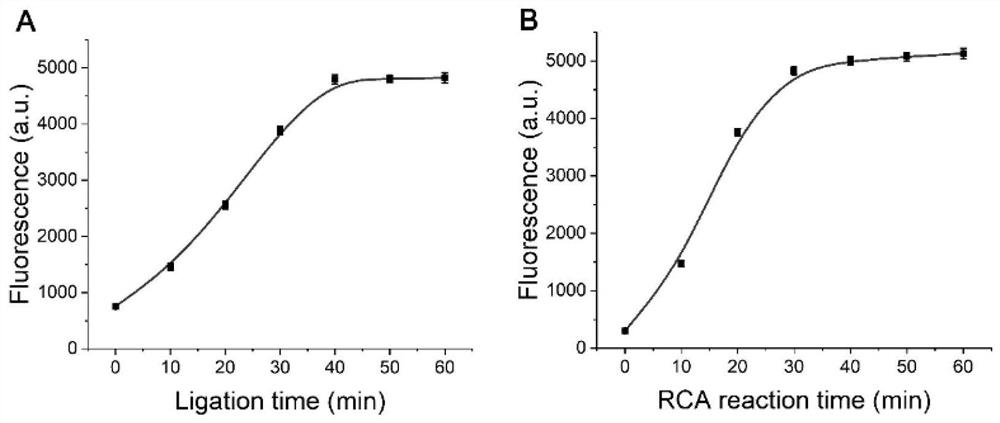
[0031] Condition optimization experiment of detection method of the present invention
[0032] The specific method is as follows:
[0033] 1) Mix 10 μL of the designed dumbbell-shaped DNA probe at a concentration of 10 nM with a 5’ end sequence fragment, 10 μL of the test sample and 30 μL of reaction buffer, and incubate at 35°C-40°C for 20-40min. Then, 1 μL, 5 U / μL T4 DNA ligase and 1× reaction buffer were added to the solution, and incubated at 22° C. for 40 min. The reaction buffer is: 15mMKCl, 30mM Tris-HCl, 15mM (NH 4 ) 2 SO 4 , 3mM MgSO 4 , pH 8.8. 10 μL of the test sample is a solution that may contain the target substance, and the solvent is water or buffer.
[0034] 2) Drop primers and 5 μL of RCA mixture (2U phi29 DNA polymerase, 1 mM dNTPs, 1×phi29 reaction buffer and 1×SGI) into the sample obtained in 1) to start the rolling circle amplification RCA reaction. The primer sequences and template sequences in 1) are shown in the table below:
[0035] Table 1 DNA...
Embodiment 2
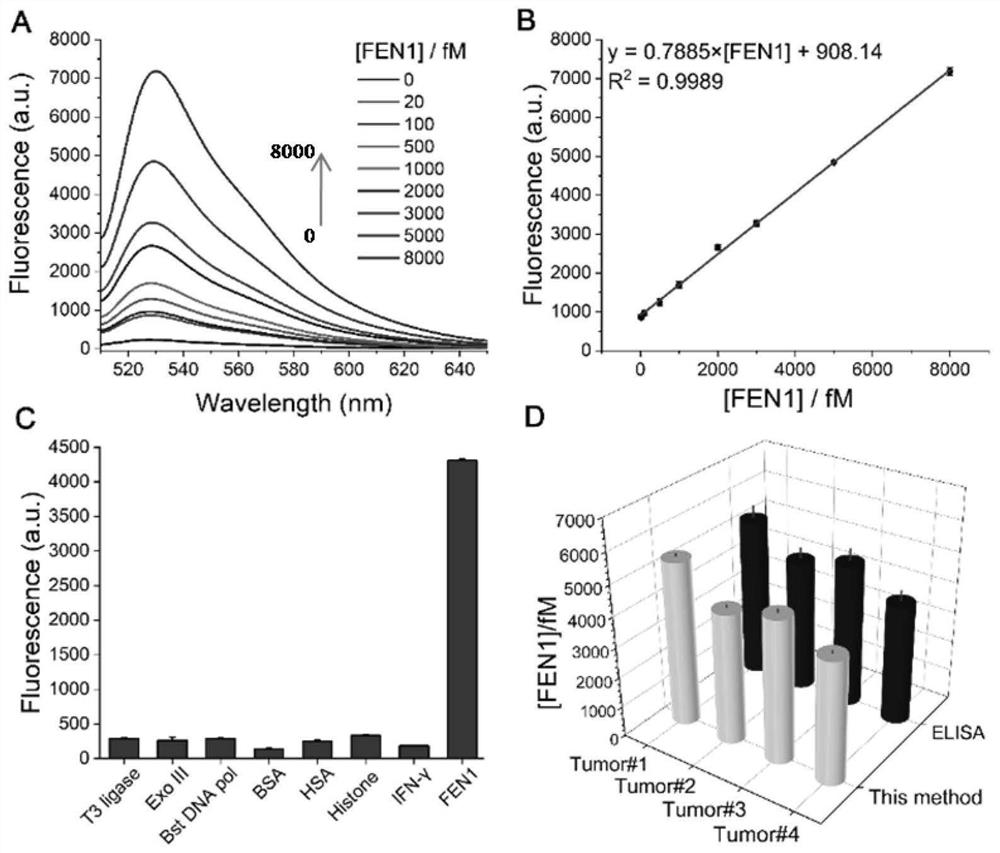
[0041] The test sample in step 1) in Example 1 was replaced with different concentrations of FEN1, and other conditions remained unchanged, and steps 1)-3) in Example 1 were repeated to detect fluorescence. Among them, the specific preparation method of different concentrations of FEN1 is: dilute the FEN1 standard with reaction buffer to obtain FEN1 solutions with concentrations of 0, 20, 100, 500, 1000, 2000, 3000, 5000, and 8000 fM.
[0042]Draw the fluorescence spectrum of the sensing system corresponding to different concentrations of FEN1, such as image 3 As shown in A. Plot the linear relationship between FEN1 concentration and fluorescence signal as image 3 Shown in B. According to linear fitting, the obtained linear range is 20-8000 fM, and according to the calculation rule of signal-to-noise ratio=3, the detection limit is 15 fM.
Embodiment 3
[0044] Selective experiment of detection method of the present invention
[0045] Replace the test sample in step 1) in Example 1 with corresponding concentrations of T3 DNA ligase, Exo III, Bst DNA pol, BSA, HAS, histone and IFN-γ, and repeat the steps in Example 1 with other conditions unchanged 1)-3), detect fluorescence, and obtain the selective result of detecting target FEN1 by the method of the present invention, such as image 3 C shown. from image 3 In C, it can be seen that the method of the present invention has good selectivity to the target FEN1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com