Primers and method for rapidly identifying and quantifying schizosaccharomyces pombe
A technology of Schizosaccharomyces pombe and yeast, which is applied in the field of bioengineering, can solve the problems of low success rate, long cycle and high cost, and achieve the effects of high accuracy, good effectiveness and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Design of Schizosaccharomyces pombe specific primers
[0034] Design method of specific primer pair:
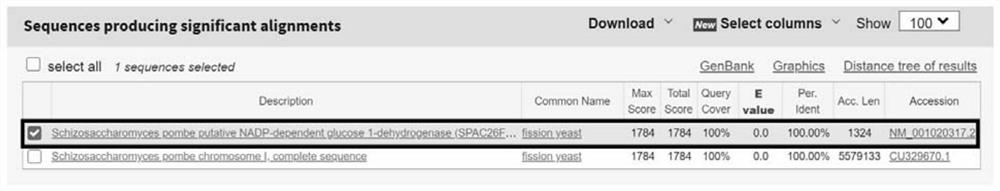
[0035] Schizosaccharomyces pombe is the core yeast in the liquor brewing system, and the Kyoto Encyclopedia of Genes and Genomes Metabolic Pathway (KEGG Pathway) was used to search for the specific enzyme of Schizosaccharomyces pombe—alcohol dehydrogenation in Schizosaccharomyces pombe Enzyme (Schizosaccharomyces pombe alcohol dehydrogenase NADP), obtain the 966bp nucleotide sequence encoding this specific enzyme, the sequence is as follows:
[0036] atgtctgctgaacaaaagtatttcgaaaacgctcaaaacgttcatttcacacttgctgatggcagcaaaatccccggtttgggattgggtacctggagatcggagcccaatcaaactaaaaatgccgtaaagactgccttacaatatggctatcgccatattgatgcagcagccatctacggtaacgaggatgaagtaggtgatggcattaaagagagtggtgttcctcgcaaggacatttgggttacctcgaagctttggtgtaatgctcacgctcccgaggcagttcccaaggctttagagaagacattgaaagacttaaagttagactacttggatgaatatcttatccattggcctgtcagctttaagactggtgaggataaattccccaaagacaaggatggaaatctca...
Embodiment 2
[0039] Example 2: Screening of Schizosaccharomyces pombe specific primers
[0040] First, the core yeast was screened from the fermentation process of Maotai-flavor liquor, and 10g of fermented grains were weighed into a sterilized 250mL Erlenmeyer flask containing 100mL of sterile PBS buffer and 3g of glass beads, and then placed at 30°C, Shake at 200r / min to mix for 30min, and let stand for 5min to obtain the bacterial suspension. Dilute the bacterial suspension, take 10 -3 、10 -4 and 10 -5 Three dilutions of sample bacterial suspensions were spread on YPD medium plates and cultured in a 30°C incubator for 72 hours. Single colonies on the plates were picked and numbered as the core yeast analogous strains.
[0041] Then, the sorghum extract used for the production of Maotai-flavor liquor was used to prepare the slant medium, and the screened core yeast similar strains were isolated and purified, then transferred to the test tube slant, cultured at 30°C, and stored at 4°C ...
Embodiment 3
[0056] Example 3: Validation of Schizosaccharomyces pombe-specific primer pairs
[0057] Verification of specific primer pairs:
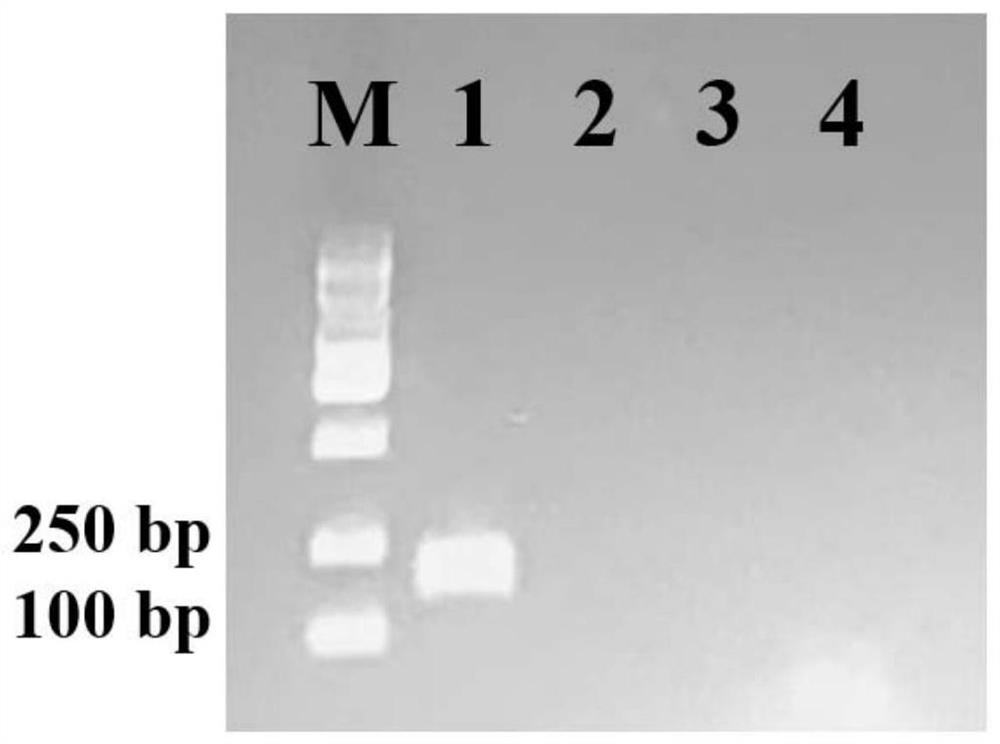
[0058] PCR was performed using the extracted genome of Schizosaccharomyces pombe (SP) as a template and the above-mentioned specific primers R1A1F-GCTGATGGCAGCAAAATCCC and R1A1R-TTGCGAGGAACACCACTCTC as primers. Negative controls were the core yeast Saccharomyces cerevisiae in the liquor brewing system: Pichia kudriavzevii (PK), Saccharomyces cerevisiae (SC), Zygosaccharomycesbailii (ZB), white sham Silk yeast (Candida humilis, CH), Pasteurian yeast (Kazachstania barnettii, KB) and redistilled water (dd H 2 O). The PCR system and PCR conditions are the same as in Example 2. After the PCR is finished, the product is subjected to gel electrophoresis, and the bands on the gel are observed by a gel imager to confirm the specificity of the primers, such as figure 2 As shown, Schizosaccharomyces pombe has obvious target bands around 100-250 bp, and th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com