A single-sample tumor DNA copy number variation detection method and device
A copy number variation, single-sample technology, applied in the field of tumor DNA copy number variation detection, can solve the problems of complicated operation, low resolution, and inability to quantify, and achieve the effect of overcoming low sensitivity and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
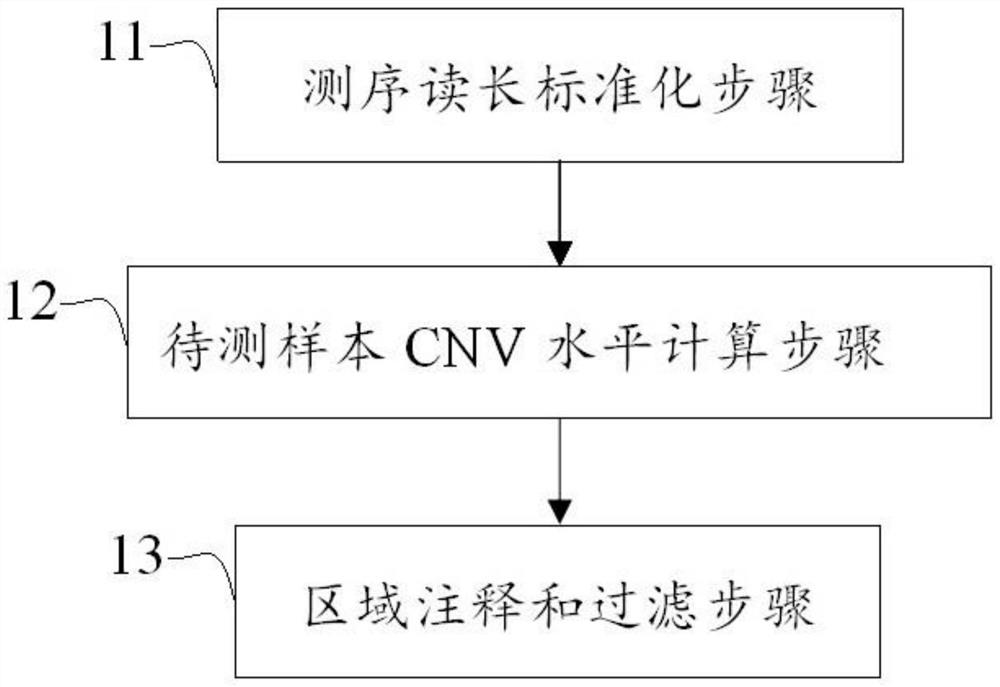
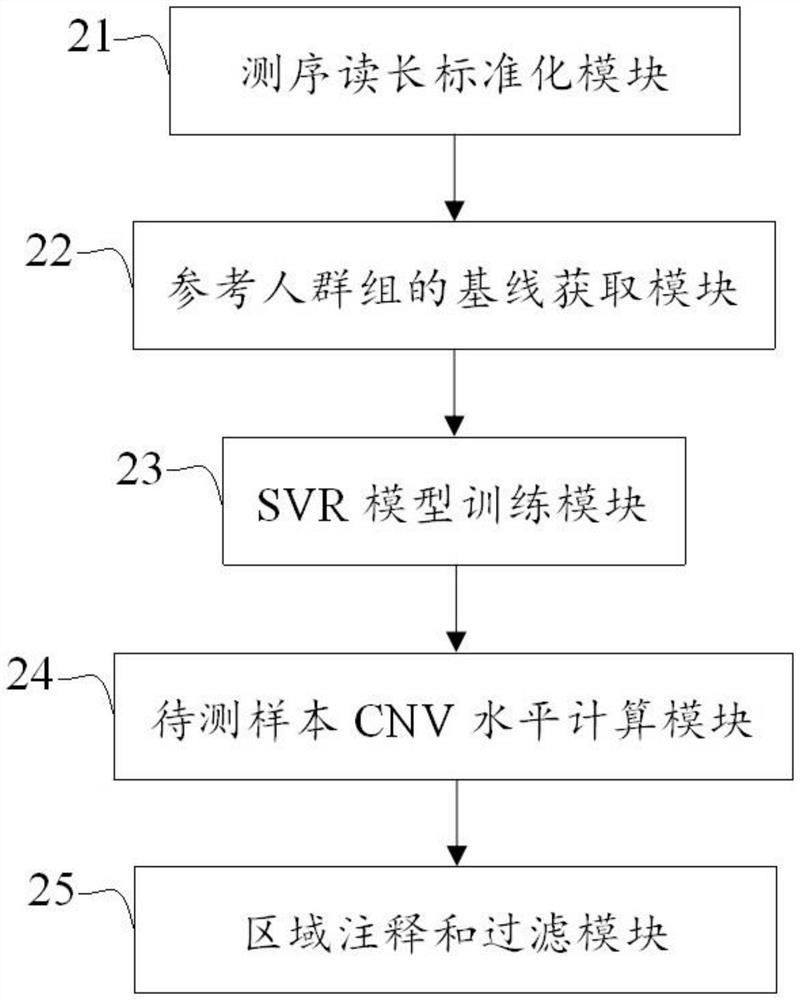
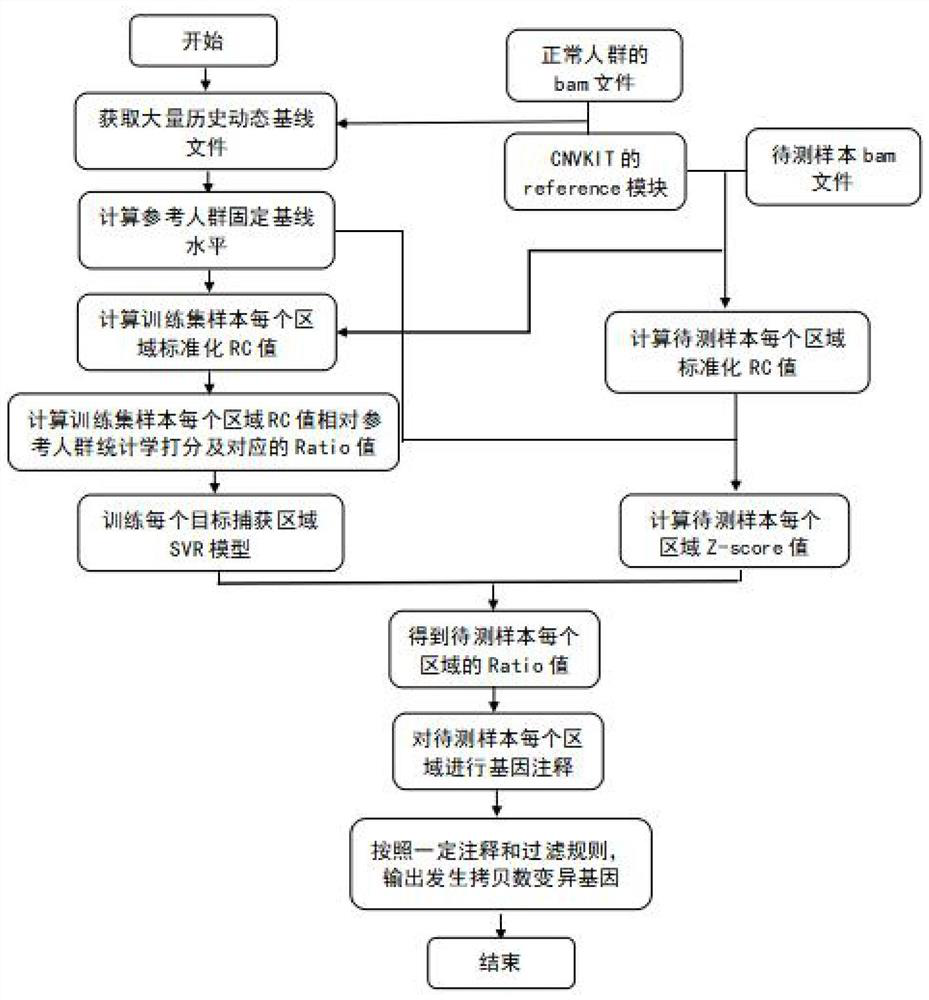
[0075] In this test, the copy numbers of MET and ERBB2 genes were identified according to the above method.
[0076] Select the 138-day historical baseline file as a reference group, calculate the mean and variance of RC in each target capture area, and use it as a fixed baseline file. Similarly, 476 clinical tissue samples were selected as the training set samples, and the CNVkit software was used to perform capture sequencing and sequencing data preprocessing, and the fixed baseline file was used as the input file to calculate the relative RC of the training set samples in each probe capture region. The statistical scoring value of the fixed baseline, that is, the Z-score value: as an indicator of the CNV level of the sample in each probe capture region, each capture region is extended back and forth by 105KB, and the target capture region within this length is used as the target capture region The characteristic value of the region and the Ratio value corresponding to the r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com