Mycobacterium tuberculosis sRNA fluorescent quantitative PCR standard substance for identifying false positive reaction and application thereof
A technique for the quantification of mycobacterium tuberculosis and fluorescence, applied in the field of microbiology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1 sRNA standard product sequence and primer design
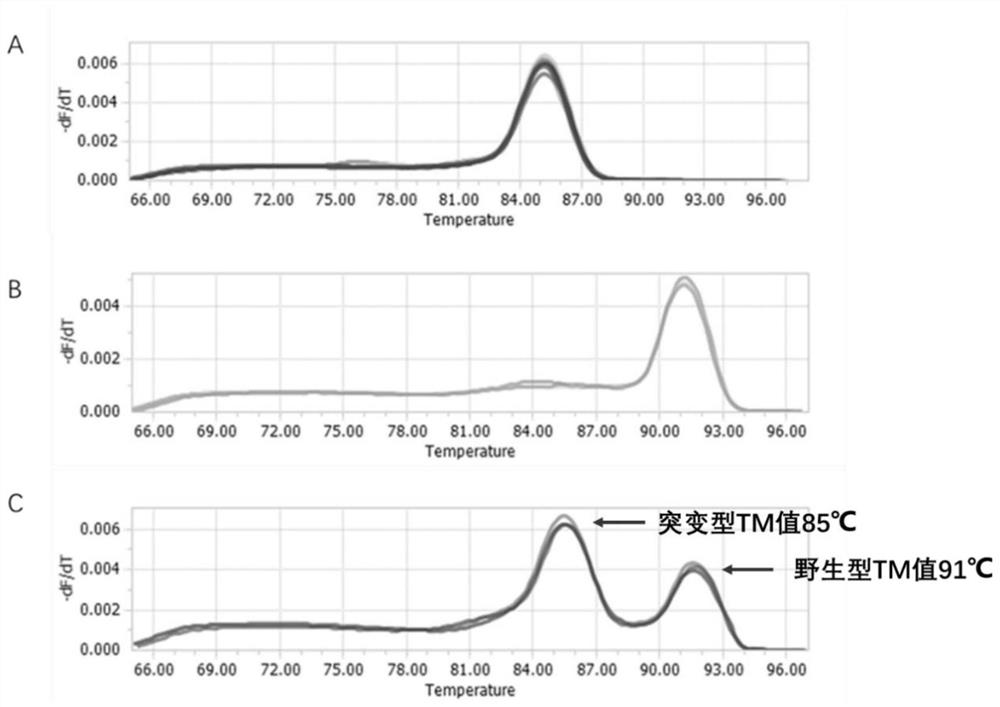
[0033] Select a wild sequence in the gene sequence of Mycobacterium tuberculosis small RNA MTS0997, and delete some bases, so that the theoretical TM value of the mutated nucleotide sequence is 4-6°C different from that of the wild sequence. Compared with the original sequence, The sRNA standard sequence with a mutation site in the middle of the amplified sequence was named #0997(-21), and was synthesized by Nanjing GenScript Biotechnology Co., Ltd. The full length of the standard is 92nt, and the PCR amplification length is 86nt.
[0034] Table 1 sRNA standard sequence
[0035]
[0036] 2.PCR primer sequence
[0037]The primers used to amplify the #0997(-21) sequence and the MTS0997 sequence in bacteria are the same set, and the primer sequences are shown in Table 2.
[0038] Table 2 Primer sequences for amplifying RNA
[0039]
Embodiment 2
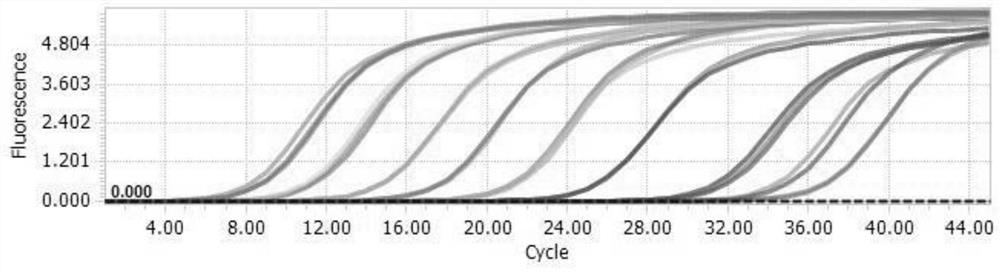
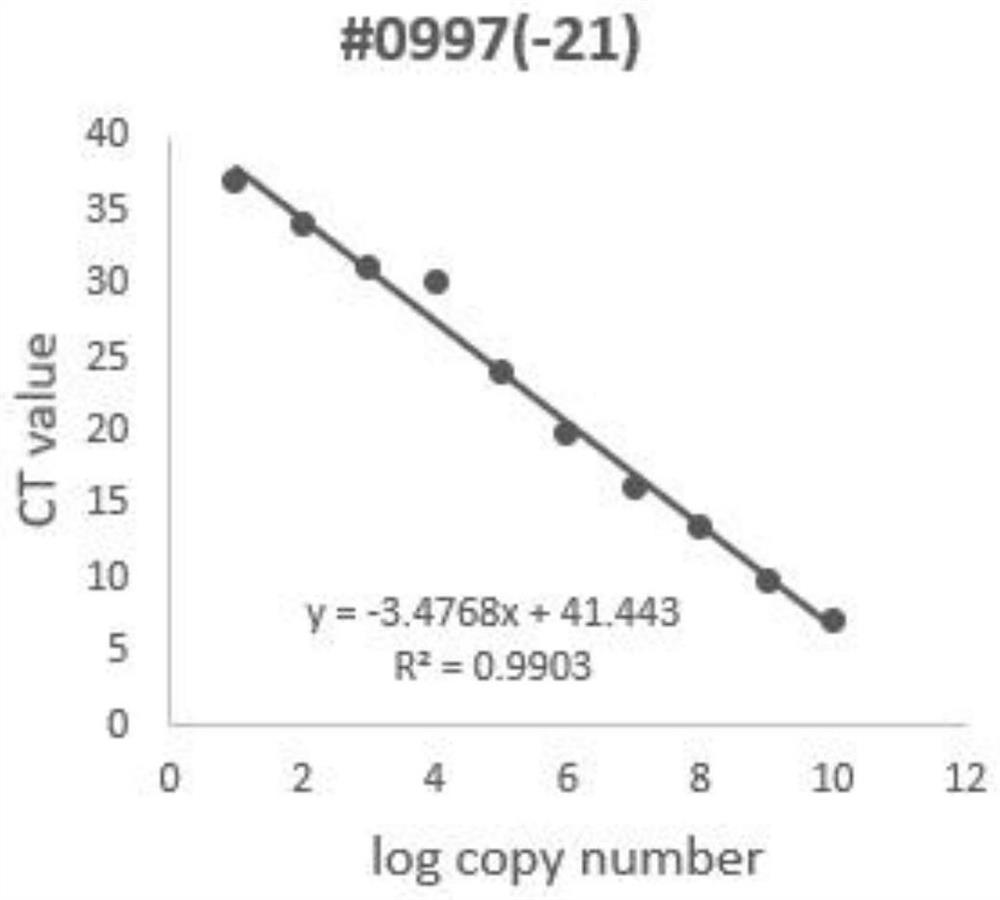
[0040] Embodiment 2 establishes sRNA standard substance real-time quantitative PCR detection system and draws standard curve
[0041] 1. RNA reverse transcription
[0042] (1) Take one tube (0.6 nmol) of sRNA standard #0997(-21) (SEQ ID No.1) dry powder and dissolve it in 100 μl DEPC-treated water. 1mol RNA is 6.02×10 23 copy number (copies), 0.6nmol RNA dry powder standard is 3.6×10 14 Copies, 100μl DEPC-treated water dissolved to 3.6×10 12 copies / μl.
[0043] (2) sRNA standard substance is carried out 10 fold dilutions (10 12 -10 0 copies / μl, 10 μl of RNA standard and 90 μl of DEPC-treated water).
[0044] (3) Take 1 μl sRNA standard of each concentration for reverse transcription.
[0045] Reverse transcription system:
[0046]
[0047]
[0048] The reverse transcription program was: 10 minutes at 25°C, 15 minutes at 42°C, and 5 minutes at 85°C.
[0049] 2. Take 2 μl of cDNA for each dilution factor for qPCR experiment
[0050] (1) qPCR system (including pri...
Embodiment 3
[0059] Stability and repeatability test of embodiment 3 sRNA standard substance
[0060] The standard curve making process of the sRNA standard in Example 2 was repeated three times. The CT values of the three results are shown in Table 3. The experimental results showed that the CVs obtained in three repeated experiments were all less than 1.5%, indicating that the sRNA standard had high stability and repeatability.
[0061] Table 3 Stability and repeatability of sRNA standards
[0062]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com