New functions and applications of nucleoside transferase
A transferase and nucleotide technology, applied in the field of genetic engineering, can solve the problems of low DNA polymerase activity and difficult enzymatic oligonucleotide synthesis, and achieve high substrate specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1 Obtaining of TDT protein
[0065] 1. Amino acid sequences of TDT proteins from different species
[0066] The amino acid sequences are as follows: SEQ ID NO:1 (mouse), SEQ ID NO:2 (platypus), SEQ ID NO:3 (myotis), SEQ ID NO:4 (naked zokor), SEQ ID NO: 5 (chimpanzee), SEQ ID NO: 6 (gecko), SEQ ID NO: 7 (emu), SEQ ID NO: 8 (junga chicken), SEQ ID NO: 9 (white-throated finch), SEQ ID NO: 10 ( Peregrine Falcon), SEQ ID NO: 11 (Golden Eagle). The homology is shown in Table 1:
[0067] Table 1 Homology analysis of amino acid sequences of TDT proteins from different species
[0068]
[0069] 2. Construction of expression vector
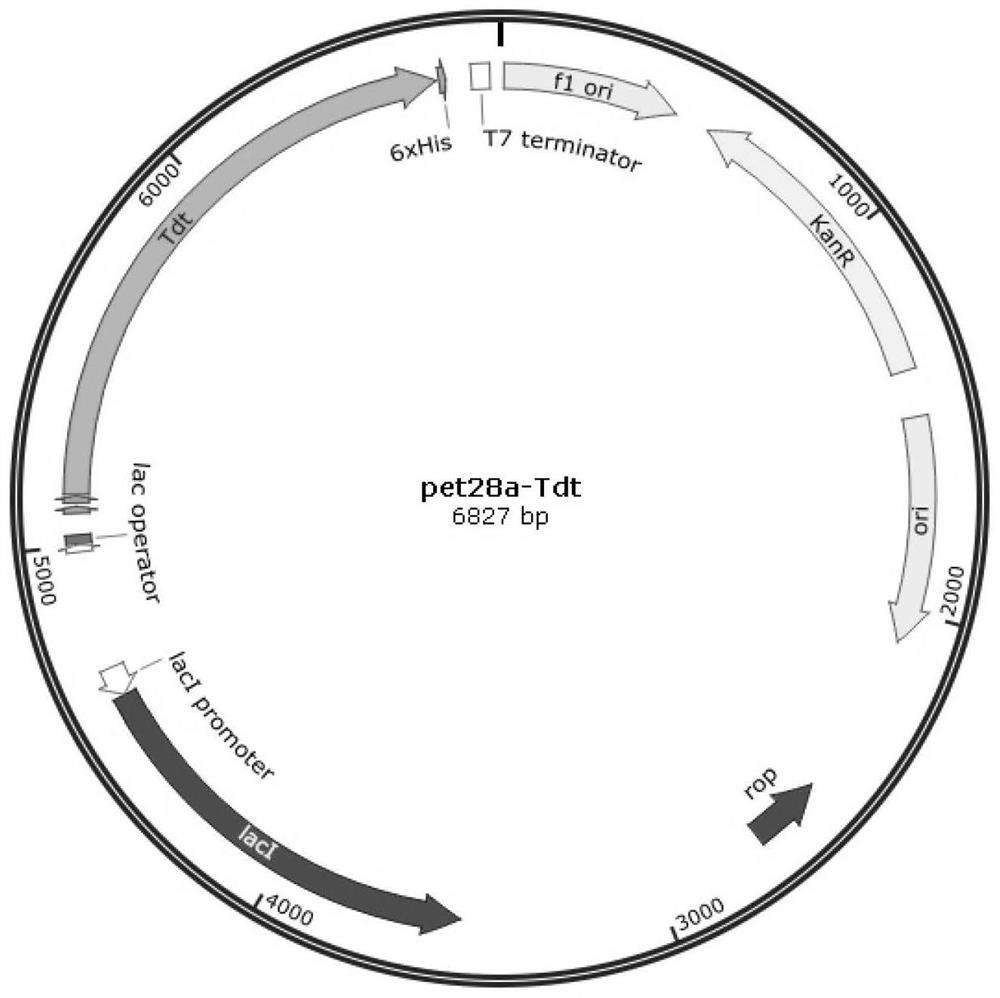
[0070] All gene sequences that can synthesize the amino acid sequences shown in the sequence listing can be used in the construction of expression vectors. In this embodiment, the gene sequence encoding TDT protein in the above-mentioned species recorded in NCBI is constructed into the expression vector pET-28a (Novagen, Kan. + ,See...
Embodiment 2
[0086] Example 2 Function Verification
[0087] The described adopting substrate of the embodiment of the present invention is:
[0088] (1) Deoxyribonucleotides (dNTPs), including adenine deoxyribonucleotides, guanine deoxyribonucleotides, cytosine deoxyribonucleotides, and thymine deoxyribonucleotides;
[0089] (2) Based on the deoxyribonucleotide in (1), the 3'-OH end is changed to a reversible blocking group, preferably, the modification group at the 3' end is an amino group.
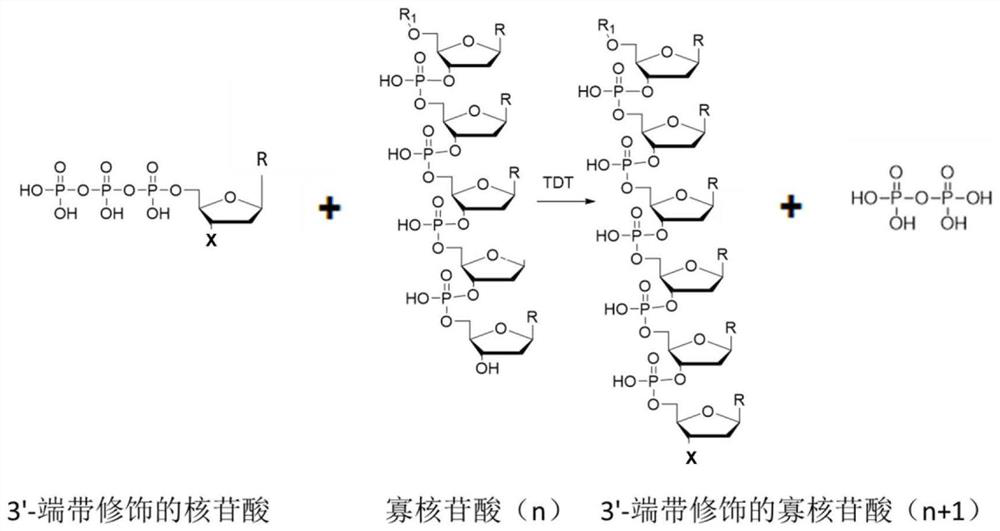
[0090] In vitro pure enzyme reaction, the reaction schematic diagram see figure 2 .
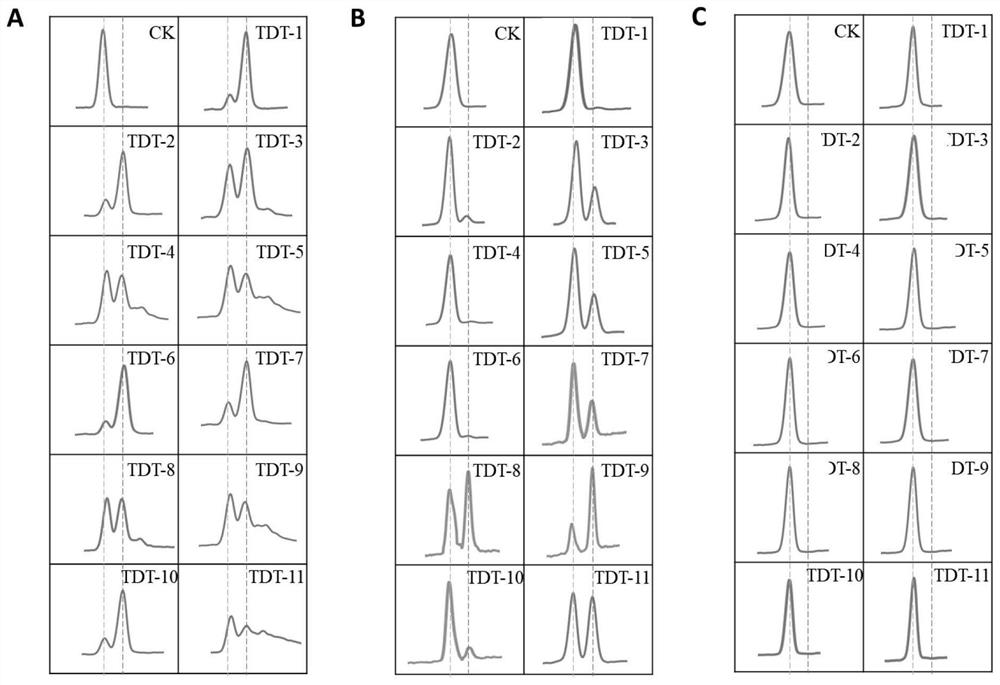
[0091] Reaction system: 100mM NaCl, 0.25mM CoCl 2 , 50mM KAc, 10mM Mg(Ac) 2 , pH 6.8. Substrate: 1 μM starting sequence 1 (14bp), 100 μM deoxyribonucleotides or 100 μM deoxyribonucleotides with amino groups at the 3’ end. Enzyme: 50 μM TDT proteins from different species prepared in Example 1. Starting sequence 1: TAATACGACTCACT
[0092] Reaction conditions: react at 30°C for 30s, inactivate the protein at...
Embodiment 3
[0094] Embodiment 3 function verification
[0095] The described adopting substrate of the embodiment of the present invention is:
[0096] (1) Deoxyribonucleotides (dNTPs), including adenine deoxyribonucleotides, guanine deoxyribonucleotides, cytosine deoxyribonucleotides, and thymine deoxyribonucleotides;
[0097] (2) Based on the deoxyribonucleotides in (1), the 3' end is changed to a reversible blocking group. Preferably, the modification group at the 3' end is an oxygen amino group; preferably, the modification group at the 3' end is an oxyallyl group; preferably, the modification group at the 3' end is an oxygen azido group; preferably, the modification group at the 3' end is an oxygen phosphoric acid group.
[0098]
[0099] The 3' deoxyribonucleotide with a reversible blocking group provided above is an example of thymine deoxyribonucleotide, and the difference between the other three deoxyribonucleotides and this example is only at the base end, 3 The reversible ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com