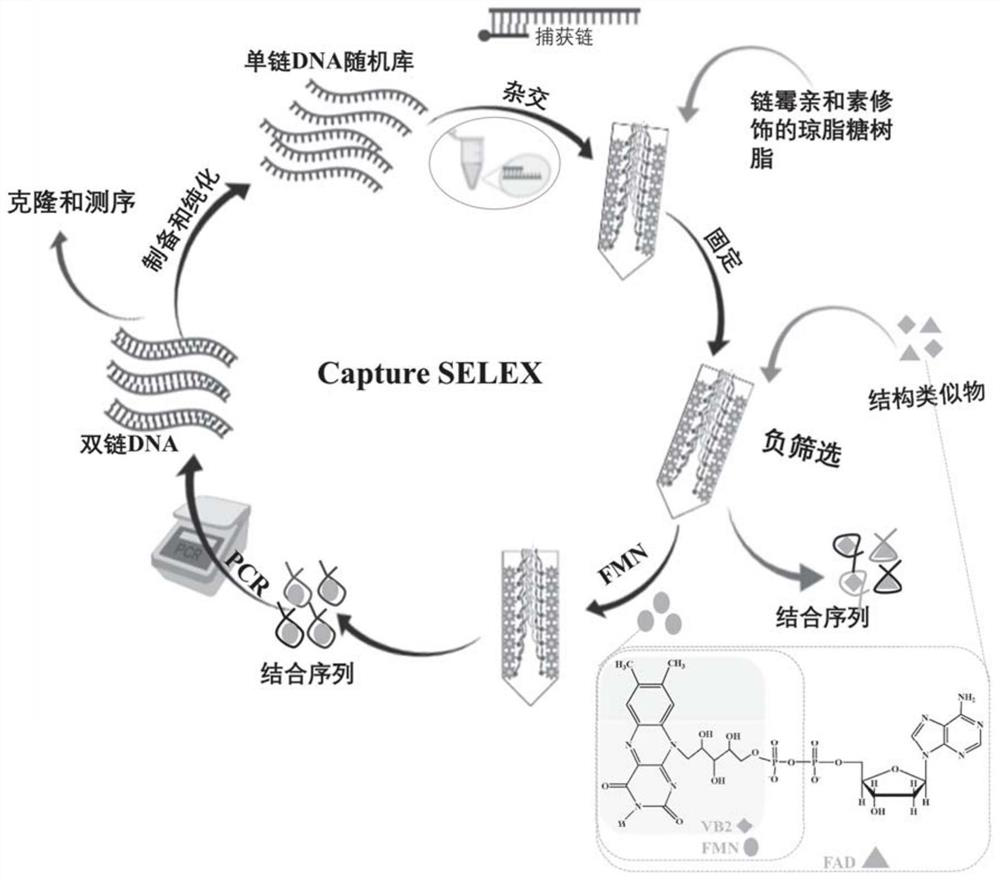
Nucleic acid aptamer of flavin mononucleotide as well as screening method and application of nucleic acid aptamer
A technology of flavin mononucleotide and nucleic acid aptamer, which is applied in chemical instruments and methods, biochemical equipment and methods, instruments, etc., and can solve problems such as poor specificity, high cost, and limited application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0102] In this embodiment, gel electrophoresis is used to monitor the screening process, and the specific steps are as follows:
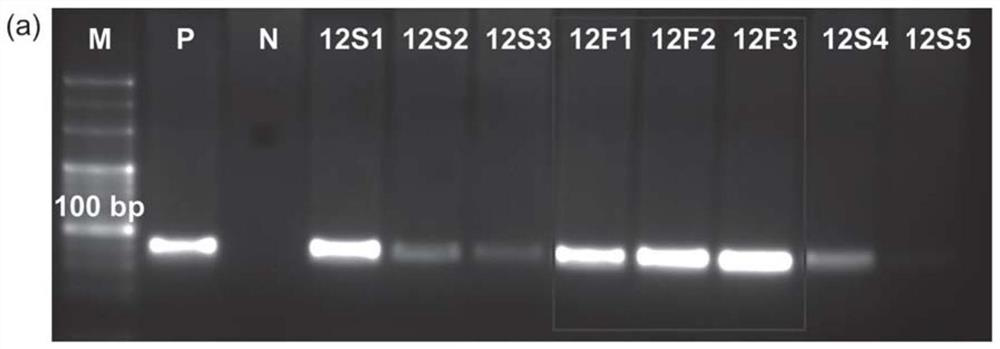
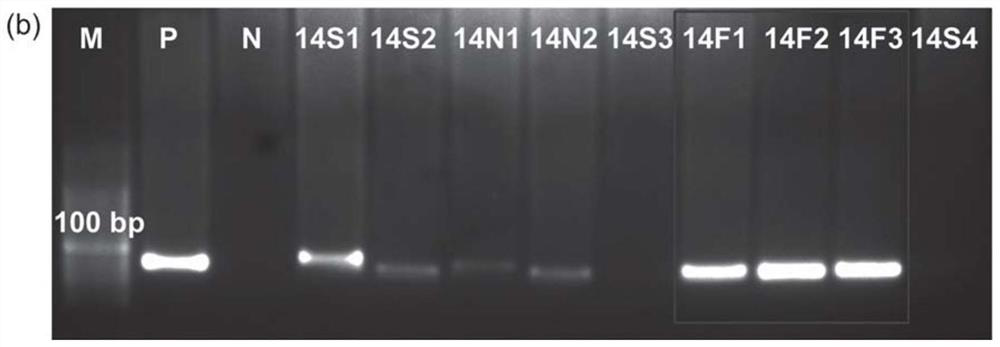
[0103] During each round of selection, the progress of SELEX was monitored using the relative amount of FMN-eluted ssDNA. The samples eluted with SELEX buffer, FMN and the mixed solution of VB2 and FAD were respectively used as templates for PCR amplification, followed by 4% agarose gel electrophoresis imaging. The result is as Figure 2a As shown, after 12 rounds of positive screening, the bands of FMN-eluted samples (12F1, 12F2, and 12F3) were significantly brighter than those of SELEX buffer-eluted samples (12S1, 12S2, 12S3, and 12S4), indicating FMN-binding sequences successfully enriched. Then, a washing step of FAD and VB2 mixed solution was introduced before positive screening, that is, negative screening to remove non-specific binding sequences. Such as Figure 2b As shown, after two rounds of negative selection, the bands of FMN eluted ...
Embodiment 2
[0105] In this example, the following methods are used to perform sequence analysis on the cloned sequencing results and select aptamer candidate sequences, and use fluorescence spectroscopy to investigate the binding ability of the selected sequences to FMN. The specific steps are as follows:
[0106] (1) After cloning and sequencing, a total of 39 sequences were obtained. The obtained sequences were compared and analyzed using the clustalx1.8.3 software, and in order to be more conducive to the selection of aptamer sequences, the fixed primers at both ends were removed, and only the part containing 30 random bases in the middle was arranged and aligned. analyze. Such as Figure 3a As shown, several sequences are repetitive, for example, sequence 1 has 6 repetitions in 40 randomly selected clones. Sequences with high repetition frequency are the preferred aptamer candidate sequences, and several aptamer candidate sequences were selected accordingly. In addition, the full-l...
Embodiment 3
[0109] In this embodiment, the affinity of the nucleic acid aptamer of the obtained FMN is tested by fluorescence spectroscopy, and the specific steps are as follows:
[0110]The aptamer was dissolved in SELEX buffer, heated at 95°C for 5min, and cooled to room temperature rapidly. Then FMN and different concentrations of aptamers were incubated in SELEX buffer at room temperature for 1 h, the final volume of the system was 100 μL, the final concentration of FMN was 1 μM, and the final concentrations of aptamers were 0, 0.1, 0.2, 0.5 , 0.75, 1, 2, 5 and 10 μM. Then use F-4600 fluorescence spectrometer to record its fluorescence spectrum in the range of 480nm-700nm, the excitation light is set to 450nm, and the slit width of excitation light and emission light is set to 10nm. The reduction value of the fluorescence intensity of FMN at 525nm is plotted against the concentration of the aptamer, using OriginPro 9.0 software and the formula (F 0 -F=F max *C / (K d +C)) for nonlin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com