Detection method and kit for lung cancer gene mutation sites
A detection kit and detection method technology, applied in the biological field, can solve problems such as reducing detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
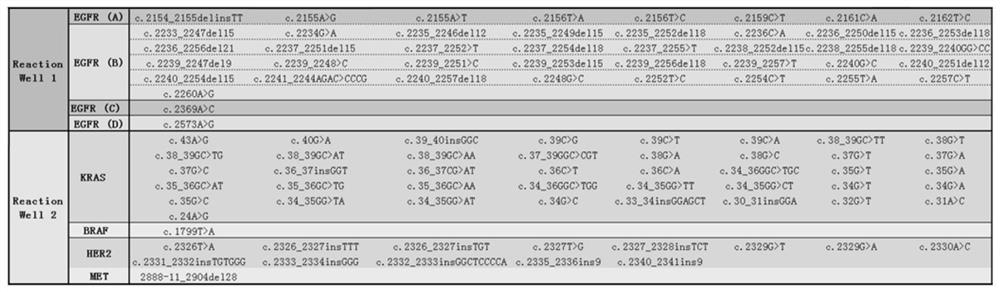
[0029] Example 1. A detection method and kit for lung cancer gene mutation sites
[0030] This embodiment mainly describes a detection method and kit for a lung cancer gene mutation site, which includes the following detection steps:
[0031] Enrolled cases: The inventor conducted a study on patients with pulmonary nodules who visited the Thoracic Surgery Department of Shanghai Zhongshan Hospital from January 2017 to December 2017. According to chest CT and blood lung cancer tumor indicators (including EGFR, KRAS, BRAF, HER2, MET ) examination results, it is impossible to determine whether it is lung cancer or benign lung disease. All patients chose to receive surgical treatment, and 10ml of peripheral venous blood was collected before the operation. According to the postoperative pathological results of the tumor, the samples were divided into lung cancer and lung benign disease groups (lung benign diseases include inflammatory pseudotumor, sclerosing hemangioma, tuberculosis...
Embodiment 2
[0073] Example 2. A detection method and kit for lung cancer gene mutation sites
[0074] This embodiment mainly describes a detection method and kit for a lung cancer gene mutation site. The difference from Example 1 is that for the primer connection nucleotide sequence shown in Table 1 and Table 2 "GGGUUGGGAAGAAACUGUGGCACUUCGGUGCCAGCAACCC (SEQ ID NO.398)", to obtain the ligated nucleotide sequence, pretreat the ligated nucleotide sequence, the specific method is: add the ligated nucleotide sequence to buffer B1 and heat to 65°C, Incubate for 10 minutes, then cool to 0°C, and incubate for 20 minutes to obtain the full-length nucleotide sequence for detection; among them, the components of buffer B1 are: 295mM NaC1, 3.5mM MgC1 2 , 15mM Tris (pH 7.6).
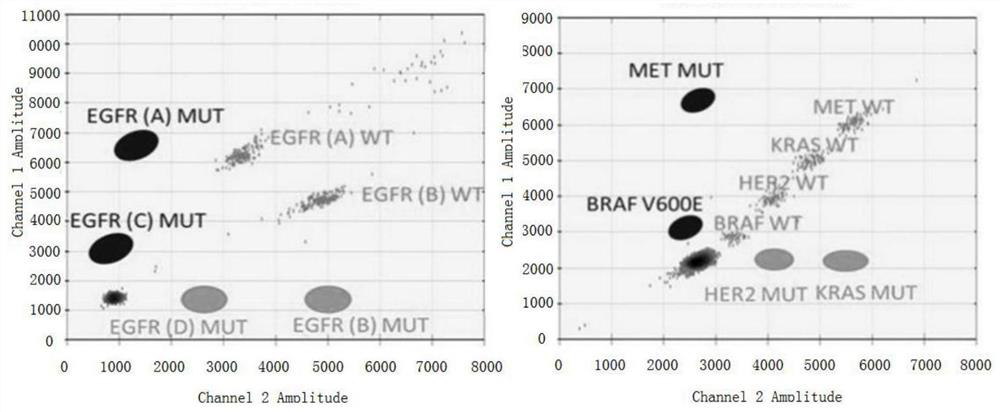
[0075] After the digital PCR amplification is completed, the effective fluorescent positive points of the two channels are interpreted by computer analysis, and the results are analyzed. Comparing this embodiment with the cons...
Embodiment 3
[0076] Example 3. A detection method and kit for lung cancer gene mutation sites
[0077] This embodiment mainly describes a detection method and kit for a lung cancer gene mutation site. The difference from Example 1 is that for the primer connection nucleotide sequence shown in Table 1 and Table 2 "GGGUUGGGAAGAAACUGUGGCACUUCGGUGCCAGCAACCC (SEQ ID NO.399)", to obtain the ligated nucleotide sequence, pretreat the ligated nucleotide sequence, the specific method is: add the ligated nucleotide sequence to buffer B1 and heat to 85°C, Incubate for 5 minutes, then cool to 40°C, and incubate for 30 minutes to obtain the full-length nucleotide sequence for detection; the components of buffer B1 are: 305mM NaC1, 5.5mM MgC1 2 , 25mM Tris (pH 7.6).
[0078]After the digital PCR amplification is completed, the effective fluorescent positive points of the two channels are interpreted by computer analysis, and the results are analyzed. By comparing this embodiment with the results of exi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com