Method and device for jointly detecting SNV, CNV and FUSEON variations
A joint detection and mutation technology, applied in biochemical equipment and methods, microbial measurement/inspection, instruments, etc., can solve problems such as complex and cumbersome operations, high cost, and low sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0194] Probe design plan for multi-gene mutation kit in blood
[0195] According to the public information of the FDA Tumor Treatment, there is an important gene such as Met, Erbb2, a MET, Erbb2, a genome range of the Erbb2 gene in a genomic range of 1MB, and select the East Asian population frequency at a genomic range of 1 Mb in a genomic range of 1 Mb. Between the SNP site between 0.3-0.7; at the same time, the frequency of 500 East Asian population is selected between 0.4-0.6, and the GC content is between 0.4-0.6, and between the crowd Highly stable SNP site. For the selected SNP site and Met, ERBB2 exon range design capture probe, the probe length is 120NT, and the target area maintains two layers of probe coverage.
[0196] According to the Open information of the FDA Tumor Treatment Division, there is an important gene such as EGFR, MET, KRAS, NRAS, BRAF, Erbb2, ALK, KIT, TP53, RB1, which are selected for the use of pharmaceutical destination design capture. The probe, the...
Embodiment 2
[0200] The construction of the multi-gene mutation joint inspection kit in the blood
[0201] 1. End repair reaction liquid formulation
[0202] First remove the following reagents from the refrigerator - 20 ° C, and the oscillation is mixed and mixed, and a single sample formulation is shown in Table 1.
[0203] Table 1:
[0204] volume cfDNA 25µL 10X terminal repair enzyme buffer 3µL Terminal repair enzyme 1.5µL total capacity 29.5µL
[0205] 2. End repair reaction
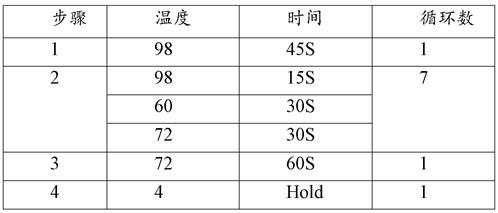
[0206] After adding 4.5 ul to the 200ul centrifuge tube, it will respond according to the proceedings of Table 2.
[0207] Table 2:
[0208] step temperature time 1 20℃ 30min 2 4℃ ∞
[0209] 3. Connect 1 Reaction MIX formulation
[0210] First remove the following reagents from the refrigerator - 20 ° C, and the oscillation is mixed and mixed, and the amount of a single sample is prepared in Table 3.
[0211] table 3:
[0212] volume Connec...
Embodiment 3
[0286] Multi-gene mutation detection in CFDNA samples in cell line mixed simulation
[0287] The cell line containing the EGFR 19DEL mutation contains KRAS G12D mutation cell line, a cell line containing EML4-Alk fusion mutation, a cell line containing MET amplification, analog CFDNA, diluted with a negative cell line, so that EGFR 19dl, Kras G12D, EML4-ALK target mutation abundance is 0.4%, 0.2% and 0.1% respectively, so that the absolute copy number of MET amplification is at 10, 6, 3.5, 2.5 copies, and utilizes Example 1. The experimental conditions of the kit and Example 2 were subjected to the sequencing sequencing data.
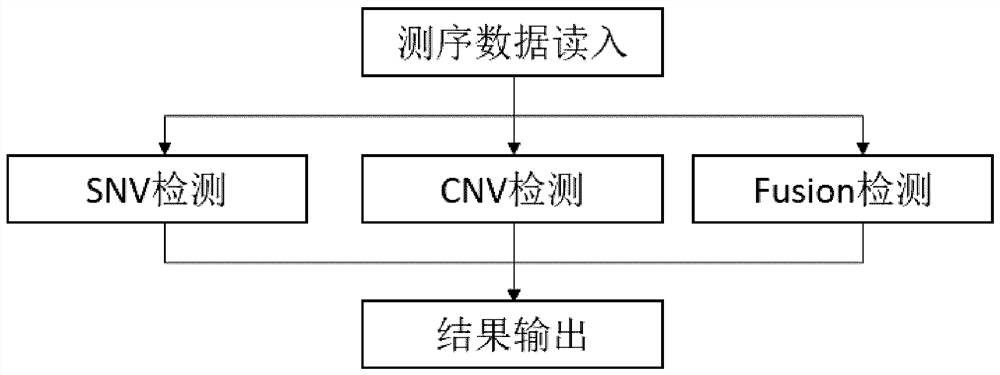
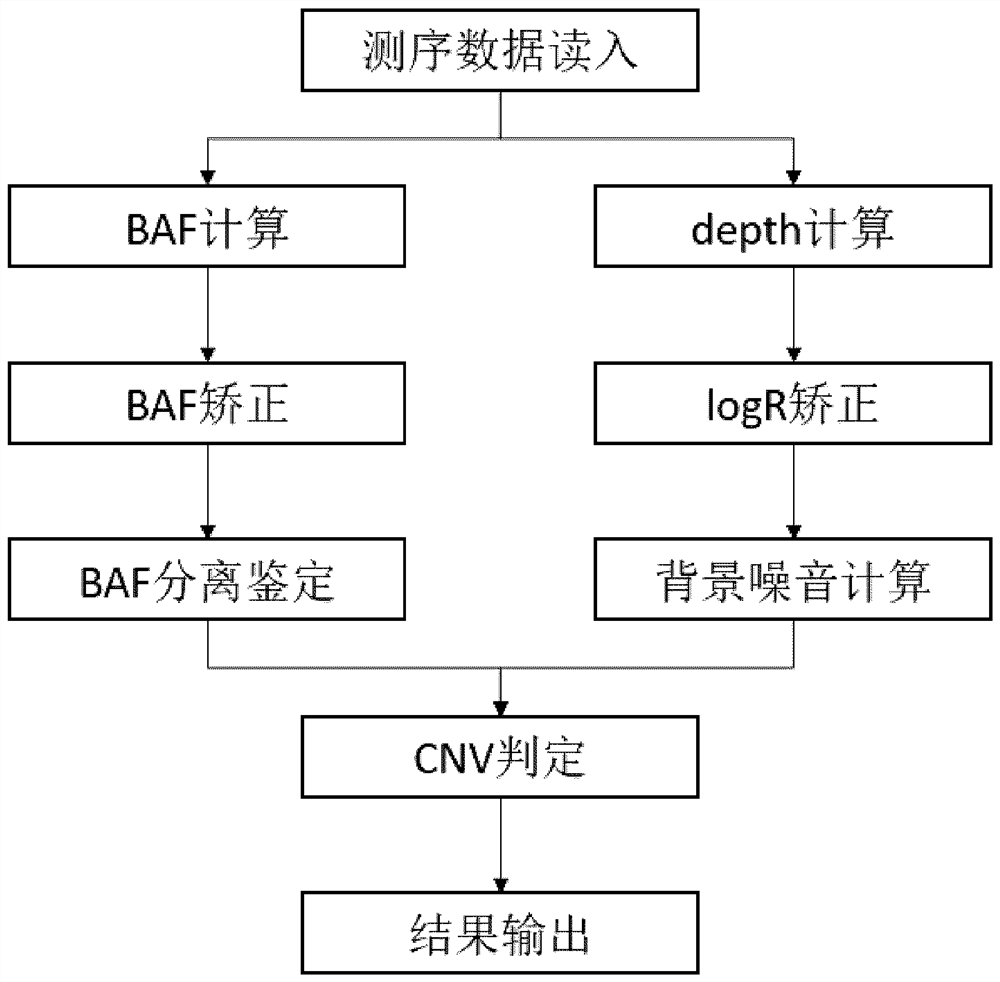
[0288] The original sequencing data generates a BAM file by BWA to the reference genome HG19, using Sambamba to order redundancy. Using conventional methods to detect SNV and Fusion mutations. CNV detection method provided by the present invention was used to detect CNV. The specific steps of CNV test are as follows:
[0289] 1) The genotype information of th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com