Construction method for physiological abundance range of N-glycopeptide of healthy people and application of construction method
A glycopeptide and physiological technology, which is applied to the construction of the physiological abundance range of N-glycopeptide in healthy people and its application field, can solve the problems of difficulty in judging the difference of N-glycopeptide and limiting the screening of tumor candidate markers.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Example 1 Constructing the physiological abundance range of N-glycopeptides in healthy people
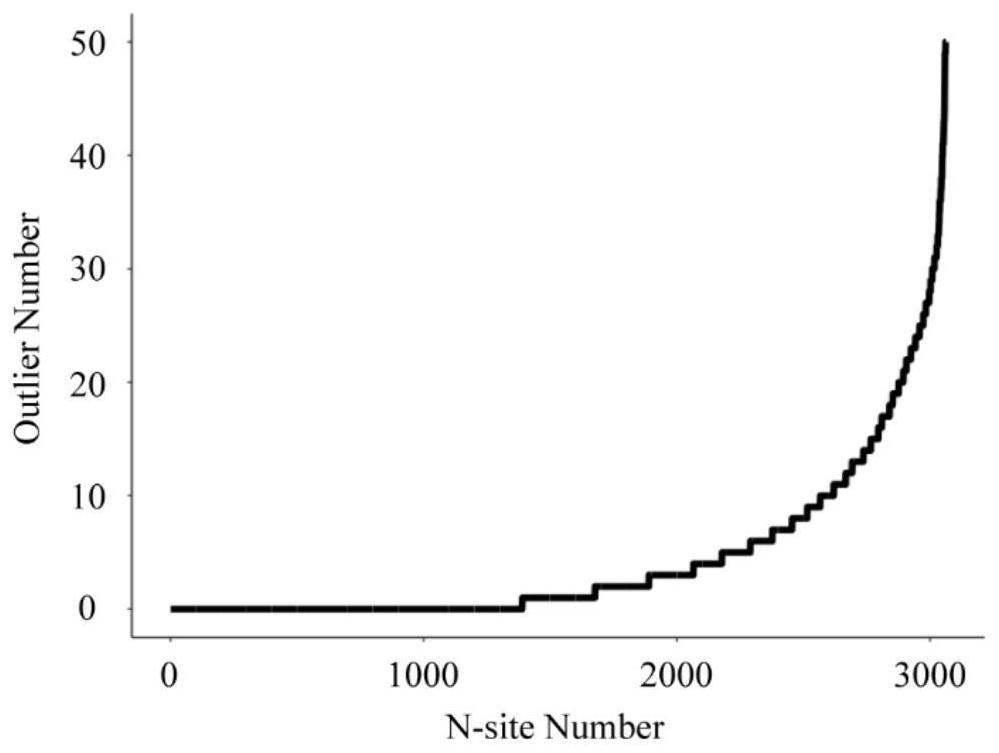
[0066] 1. Large-scale (in advance, the N-glycoproteome identification accumulation curve of 40 healthy people was investigated, in which the accumulation identification quantity of N-glycoprotein and N-glycosylation site was reached when the number of samples reached 14 cases and tends to be saturated after 20 cases) to determine the N-glycopeptide quantitative value in the physiological samples of healthy people.
[0067] Determination of the quantitative value of N-glycopeptide in physiological samples of 291 healthy volunteers, extraction of urine protein after acetone precipitation, reduction of alkylation and enzymatic digestion of peptide segment, and N-glycopeptide analysis by hydrophilic interaction chromatography (HILIC) method - Glycopeptides were enriched, N-glycan chains were cut off with peptide N-glycoamidase (PNGase F), desalted and heat-dried for mass spectrom...
Embodiment 2
[0084] Example 2 Screening for Abnormal N-Glycopeptide Potential Markers in Patients with Lung Cancer
[0085] S1. Determination of the quantitative value of N-glycopeptide in physiological samples (morning urine) of 50 lung cancer patients before treatment. The determination method refers to step 1 in Example 1.
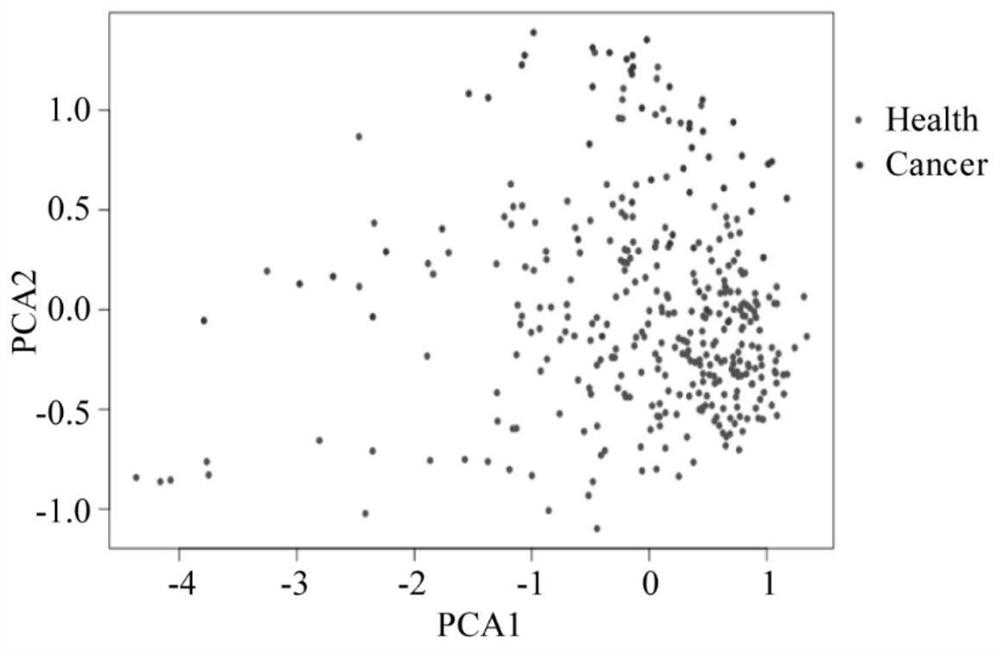
[0086] A total of 3133 N-glycopeptides were identified from the pre-treatment samples of 50 lung cancer patients and the 291 healthy human samples in Example 1. Such as figure 1 As shown, through principal component analysis, lung cancer samples can be clearly distinguished from healthy samples.
[0087] S2. Compare the quantitative value of each N-glycopeptide identified in the physiological sample of each patient with the physiological abundance range of the N-glycopeptide in the healthy population constructed in Example 1, and the N-glycopeptide whose quantitative value falls outside the range Obtaining all abnormal N-glycopeptides in the patient's physiologica...
Embodiment 3
[0104] Example 3 Screening and analysis of abnormal N-glycopeptides in 20% of lung cancer patient samples
[0105] Molecular typing research using pre-treatment samples (morning urine) of 50 patients with lung cancer: the specific steps are as follows:
[0106] Using 740 N-glycopeptides abnormally expressed in more than 20% of the total number of samples, the differences among samples of lung cancer patients were investigated to carry out molecular typing.
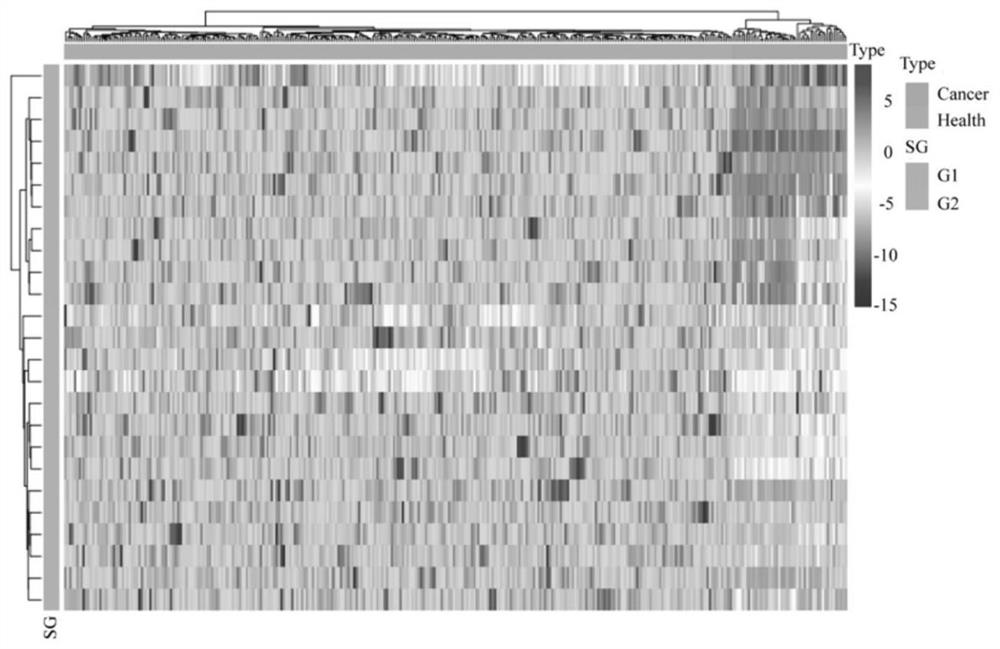
[0107] see results Figure 4 : Using a consensus clustering algorithm, these samples can be divided into four subtypes.
[0108] Figure 5 Among them, a total of 91 key N-glycopeptides were screened out from 740 N-glycopeptides, which can clearly distinguish type 1 from type 3 (C1 and C3). The column directions from left to right are respectively C1, C2, C3 and C4. The row directions from top to bottom are G6, G2, G1, G4, G3 and G5 respectively. The G1 group is mainly related to plasma lipoprotein and IGF function. I...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com