Preparation method of anti-African swine fever cloned pig
An African swine fever virus and gene technology, which is applied in the field of preparation of African swine fever-resistant cloned pigs, can solve the problems of disease-resistant breeding, lack of vaccines and treatment methods, and loss of ASF pig industry.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
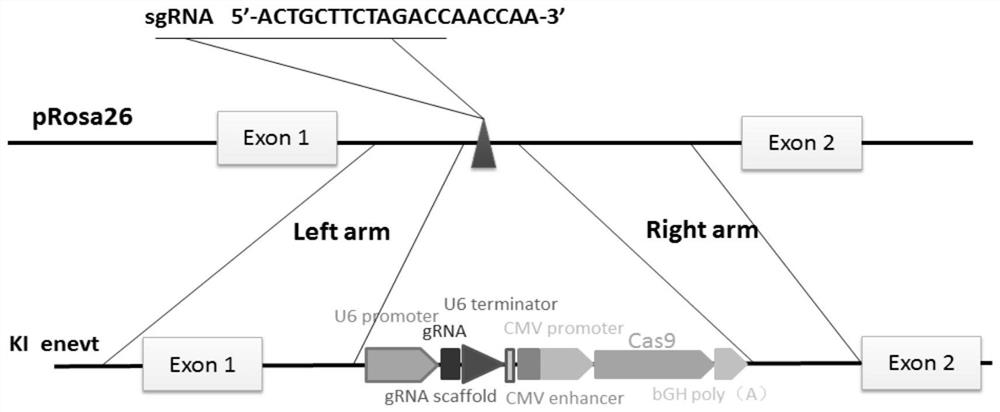
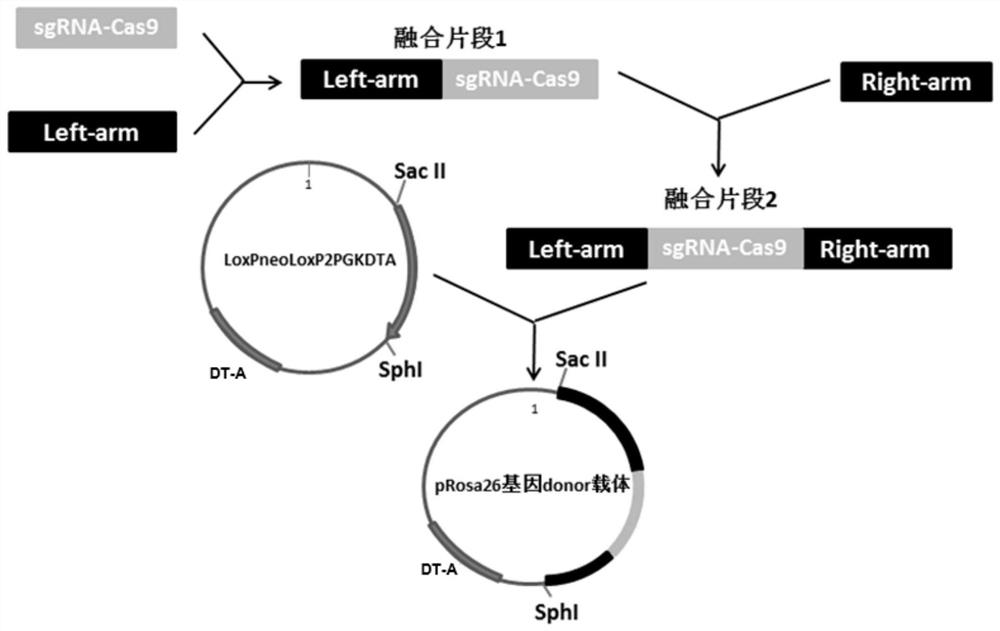
[0032] The construction of embodiment 1 pig Rosa26 gene donor carrier
[0033] S1. Fusion the sgRNA-Cas9 gene sequence capable of expressing ASFV pE248R with the homologous left arm; the sgRNA-Cas9 gene sequence was synthesized by Shanghai Sangon and verified by sequencing, and then amplified using it as a template, and the primers used for amplification (As shown in SEQ ID NO.3-4) are F5'-GAGGGCCTATTTCCCATGAT-3' and R5'-TTATCGTCCGT ACGACCCCT-3', the PCR products were purified and sequenced to verify correctness. The homologous left arm of the first intron of the porcine Rosa26 gene (as shown in SEQ ID NO.5) was amplified with 3780 bp using the porcine cell genome DNA as a template. The amplification system is: porcine cell genomic DNA, 20 μL; GreenTaq Mix, 25 μL; pRosa26-Lelf-F’ (100 μM) 2 μL; pRosa26-Lelf-R’ (100 μM) 2 μL; ddH 2 O, 1 μL. The amplification program is: 95°C, 3min; 95°C, 15sec, 60°C, 15sec, 72°C, 4min, 35 cycles; 72°C, 5min. The primer pair used for amplific...
Embodiment 2
[0036] Example 2 Construction of CRISPR-Cas9 Targeting Vector
[0037] S1. Use the online design website (https: / / benchling.com / ) to predict the targeting site of the first intron of the pig Rosa26 gene; according to the self-assessment and the scores in the prediction results, the candidate target sites are clocked The one with the highest score is selected, and its sgRNA sequence (as shown in SEQ ID NO.12) is ACTGCTTCTAGACCAACCAA. Complementary paired oligonucleotides were synthesized according to the sgRNA sequence, as shown in Table 1, where the lowercase letters are restriction sites.
[0038] Table 1 Oligonucleotide sequence
[0039] name sequence (5'-3') px459-F (as shown in SEQ ID NO.13) caccGACTGCTTCTAGACCAACCAA px459-R (as shown in SEQ ID NO.14) aaacTTGGTTGGTCTAGAAGCAGTCAGTC
[0040] S2. Construction of targeting vector
[0041] Using the oligonucleotides in Table 1, the construction process is: 94°C, 5min, then 35°C, 10min, and then ...
Embodiment 3
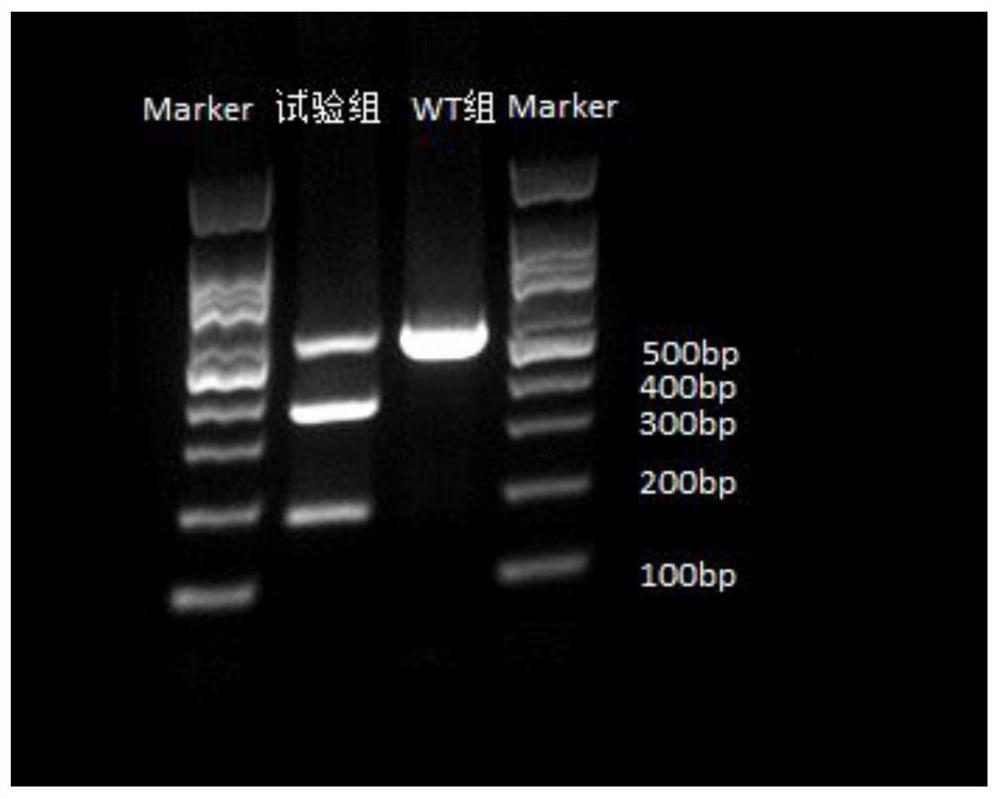
[0050] Example 3 Screening and Identification of Positive Cell Monoclonal
[0051] S1. Screening of Positive Monoclonal Cells
[0052] Digest and collect porcine fibroblasts (approximately 1 × 10 6 1), the targeting vector px459 constructed in Example 2 and the pig Rosa26 gene donor vector constructed in Example 1 were mixed according to the ratio of the amount of substances 1:2, and the total mass was 6 μg, according to the method of S5 in Example 2 After transfection, place in CO 2 In the incubator, cultivate at 37°C. After 48 hours, the confluence of the cells reached 80%-90%. At this time, the cells in one well were evenly divided into eight 10cm culture dishes. After 24 days, the cells adhered to the wall, and the medium was replaced with the fibroblast medium containing G418 (500 μg / mL), and the medium was changed every 3 to 4 days, and the medium was still the fibroblast medium containing G418 (500 μg / mL) . After the cells were cultured for 7-10 days, the formation...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com